Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Antinuclear antibody (ANA) immunofluorescence (IFA) patterns and titers are a key part of rheumatology diagnostics, however, there is considerable intra- and inter-laboratory variability with manual interpretation. Replacing manual interpretation with a standardized and automated approach could help reduce variability, increasing laboratory accuracy and efficiency. We developed machine learning (ML) models to aid laboratories in ANA pattern and titer interpretation, including a model for the nuclear dense fine-speckled (DFS) ANA pattern (AC-2), a rare pattern among systemic autoimmune rheumatic disease (SARD) patients that decreases the likelihood of these conditions.
Methods: 13,671 ANA images from SLE patients enrolled in the Systemic Lupus International Collaborating Clinics Inception Cohort (SLICC, n=2,825 images), non-SLE subjects enrolled in the Ontario Health Study (OHS, n=10,639 images), and the International Consensus on ANA Patterns (ICAP, n=207 images) were analyzed. All SLICC and OHS ANA were performed in one central laboratory using IFA on HEp-2 cells (NovaLite, Werfen, SD) and read on a digital IFA microscope (NovaView, Werfen, SD). As the reference standard, one laboratory technologist (HH) with >30 years of experience in ANA studies interpreted ANA patterns and titers for each image. We developed and compared the performance of eight ML models for 13 ANA pattern recognition. Four convolutional neural network (CNN) models and an image feature extractor were developed to differentiate AC-2 from AC-4 (speckled) and AC-30 (nuclear speckled with mitotic plate staining) patterns, which appear similar but are associated with SARDs. To evaluate ANA titer, we used an ML technique for imaging processing that identified individual HEp-2 cells in the ANA images and then calculated the cell illuminance and cut-offs corresponding to each titer (1:80-1:5120). Fifty images were randomly selected to compare the titer classification based on image processing with the lab technologist as the reference standard.
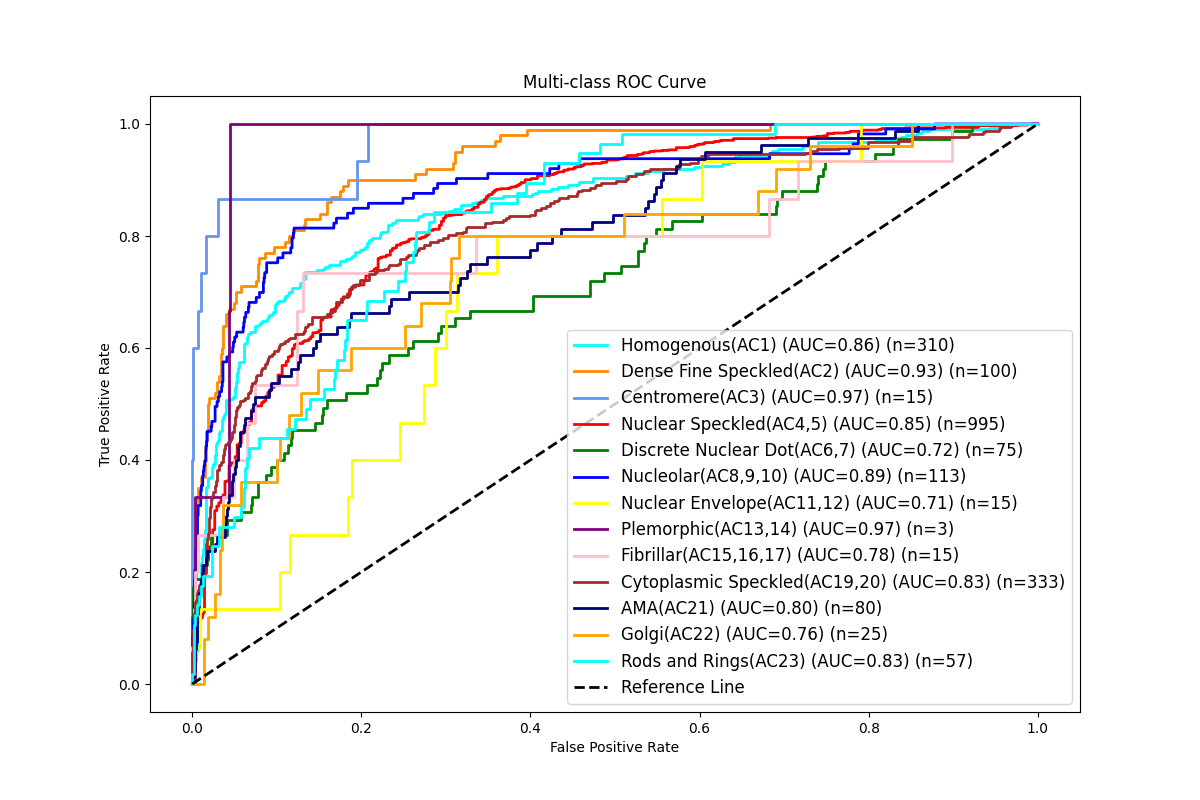
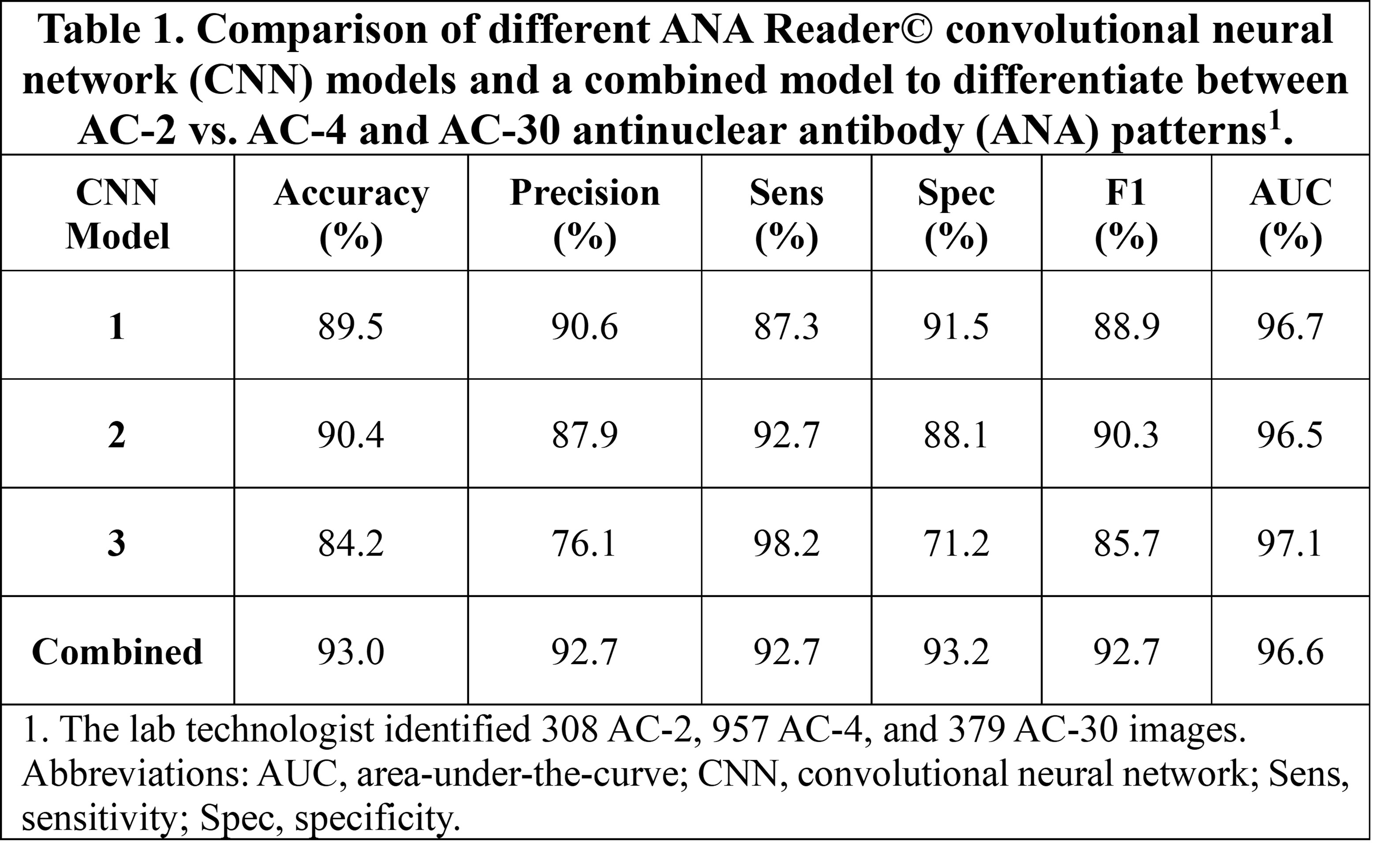
Results: We identified one ML model with the best performance for ANA pattern identification compared to the reference with a high area under the curve (AUC) score of 83.4% and more modest performance in other metrics (Figure 1). For the AC-2 models, all four CNNs performed similarly with high AUC scores (96.5%-97.1%) (Table 1). There was a strong correlation between titers reported by the identified model and the technologist’s interpretation (Spearman rank 0.93, p < 0.0001), where the titers reported were identical or differed by only one dilution in most cases (96.0%).
Conclusion: ML has the potential to become a highly effective and efficient approach to evaluating ANA patterns and titers. The performance of our model is expected to improve as we continue to train our models with more images. The AC-2 model can already discriminate the nuclear DFS pattern from other SARD-related patterns with a high degree of accuracy, potentially speeding up the differentiation of those at risk vs. not at risk of SARDs. Future external validation studies and ML models to predict more complex and multiple ANA patterns and titers are underway.
To cite this abstract in AMA style:
Choi M, Moghaddam F, Sajadi M, Clarke A, Bernatsky S, Costenbader K, Chen I, Urowitz M, Hanly J, Gordon C, Bae S, Romero-Diaz J, Sanchez-Guerrero J, Wallace D, Isenberg D, Rahman A, Merrill J, Fortin P, Gladman D, Bruce I, Petri M, Ginzler E, Dooley M, Ramsey-Goldman R, Manzi S, Jönsen A, Alarcón G, Van Vollenhoven R, Aranow C, Mackay M, Ruiz-Irastorza G, Lim S, Inanç M, Kalunian K, Jacobsen S, Peschken C, Kamen D, Askanase A, Fritzler M, Aminghafari M. Rheumatology Diagnostics Utilizing Artificial Intelligence (ANA Reader©) for ANA Pattern Identification and Titer Quantification [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/rheumatology-diagnostics-utilizing-artificial-intelligence-ana-reader-for-ana-pattern-identification-and-titer-quantification/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rheumatology-diagnostics-utilizing-artificial-intelligence-ana-reader-for-ana-pattern-identification-and-titer-quantification/