Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: T-cells play a key role in the pathogenesis of autoimmune disease and in protection against cancer and infection. Consequently, there is widespread interest in characterizing T-cell states in disease tissue using single-cell RNA sequencing (scRNA-Seq). However, such data has proven challenging to interpret. scRNA-Seq has revealed a continuum of T-cell states without discrete clusters corresponding to expected subsets (e.g. Th1, Th2, and Th17). Furthermore, it is unclear how to identify T-cells that have recently been activated by antigen. Understanding the subset and activation status of T-cells in disease tissue can shed light on pathogenesis.
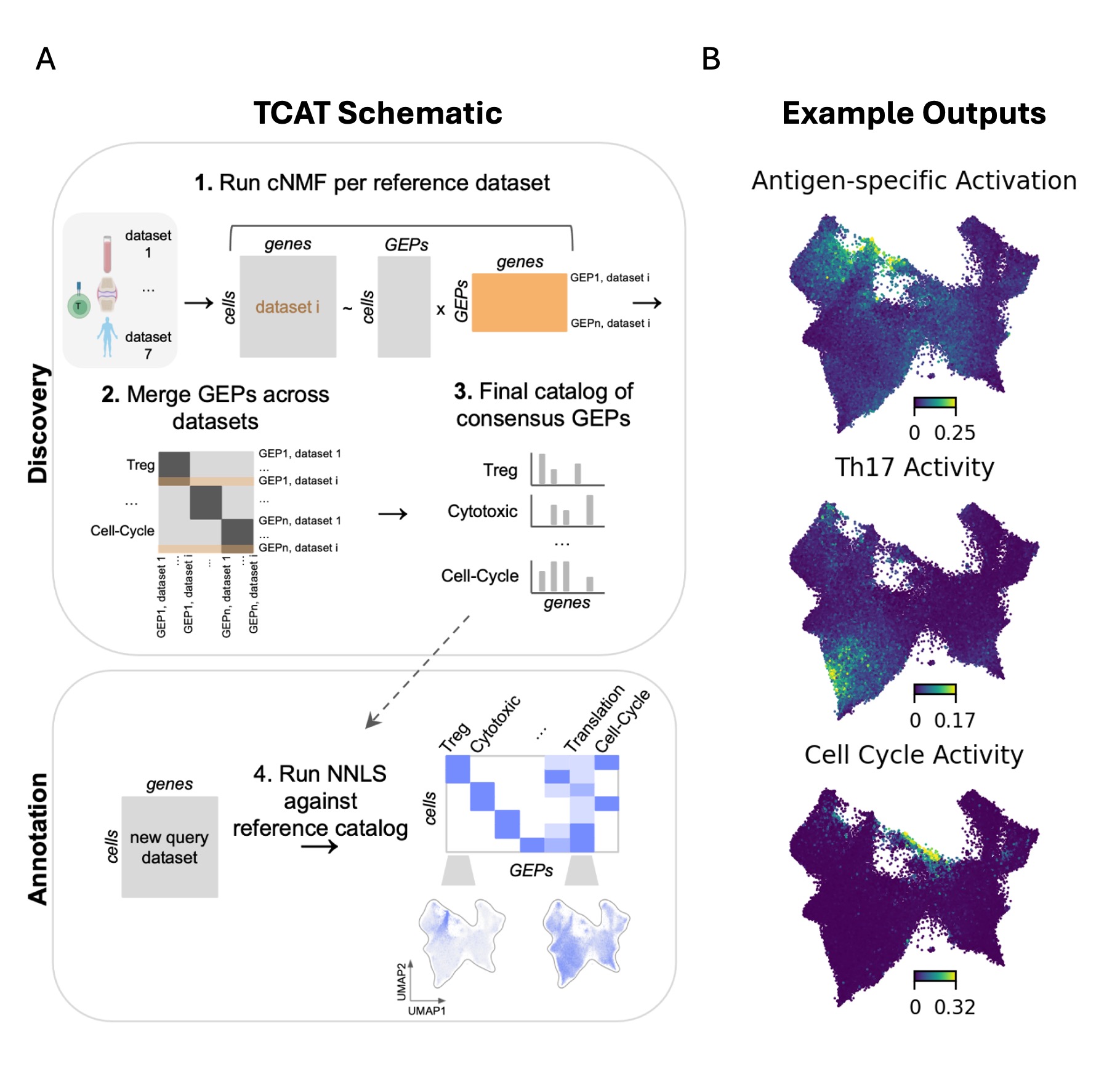
Methods: We developed T-CellAnnoTator (TCAT), a computational pipeline that overcomes these limitations of T-cell scRNA-Seq analysis. TCAT analyzes multiple reference datasets to derive a catalog of gene expression programs (GEPs) capturing the core functions of T-cells (e.g. polarization states, cytotoxicity, and exhaustion). It then infers the activity of these programs in new query datasets, enabling users to annotate subsets, antigen-specific activation status, and other GEPs in their data.
Results: We discovered 46 reproducible GEPs from 1,700,000 T-cells from 700 individuals across 38 tissues and five diverse disease contexts. These GEPs capture all known major T-cell subsets and several known and novel antigen-specific activation responses. We show that TCAT predicts T-cell subsets in silico and in vivo with high accuracy. We identify associations of GEPs with disease states and highlight the enrichment of T peripheral helper (Tph) cells in tumors and Covid-19. We show widespread Th2-polarized CD8 T-cells in peripheral blood. We also document wide variability in the percentage of activated, exhausted, and proliferating T-cells in Rheumatoid Arthritis synovia and in tumors.
Conclusion: Our software enables users to easily annotate single-cell profiles of T-cells with subsets and activation-state information, providing insight into which T-cell are being activated by antigens and which are “bystanders” within disease tissue. The overall strategy can be generalized to all cell types and tissues. We make TCAT publicly available and have created a repository to host GEP catalogs from new tissues, enabling easy application to new datasets.
Disclosures: D. Kotliar: None; M. Curtis: None; R. Agnew: None; K. Weinand: None; A. Nathan: None; Y. Baglaenko: None; Y. Zhao: None; P. Sabeti: Danaher Corporation, 11, Delve Bio, 2, 4, 11, Sherlock Biosciences, 2, 4, 11; D. Rao: Amgen, 6, AnaptysBio, 2, AstraZeneca, 1, Bristol-Myers Squibb, 2, 5, GlaxoSmithKline, 2, HiFiBio, 2, Janssen, 5, Merck, 5, Scipher Medicine, 2; S. Raychaudhuri: Janssen, 1, Mestag, 8, Nimbus, 2, Pfizer, 1, Sonoma, 8, Third Rock Ventures, 2.
To cite this abstract in AMA style:
Kotliar D, Curtis M, Agnew R, Weinand K, Nathan A, Baglaenko Y, Zhao Y, Sabeti P, Rao D, Raychaudhuri S. Reproducible Single Cell Annotation of Programs Underlying T-cell Subsets, Activation States, and Functions [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/reproducible-single-cell-annotation-of-programs-underlying-t-cell-subsets-activation-states-and-functions/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/reproducible-single-cell-annotation-of-programs-underlying-t-cell-subsets-activation-states-and-functions/