Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Citrullination is a key physiological process that drives autoantibody formation in Rheumatoid Arthritis (RA). This irreversible post-translational modification of the amino acid arginine is catalyzed by Peptidyl Arginine Deaminase (PAD), which converts the positively charged arginine residue to a negatively charged citrulline. Most RA patients develop a spectrum of autoantibodies to peptides/proteins featuring citrulline residues (ACPA). It remains unclear whether there is an excess production of specific citrullinated proteins in RA patients that is driving the autoantibody response. The aim of this study was to undertake an unsupervised proteomic analysis of the RA citrullinome, and determine the consequences of citrullination in RA.
Methods: To isolate citrullinated proteins, we used a biotinylated Phenylglyoxal probe (Cayman), which has been shown previously to bind to citrulline with high specificity. Citrullinated serum proteins were isolated on streptavidin beads from 20 samples, 10 ACPA+ RA patients and 10 First-Degree Relatives (FDR) of RA patients, who served as controls. Mass spectrometry was undertaken after on-bead digestion with trypsin. MS data was processed with MaxQuant to determine label free quantification, and a network analysis was performed in R. Enriched cit-proteins and cit-immune complexes (IC) were measured using custom ELISAs, which included a larger cohort of FDR (ACPA- n=31, ACPA+ n=26) and RA (n=31) from a well-established RA risk longitudinal cohort in Manitoba, Canada. Classical complement activation was measured by absorbance (Svar) using 10% human serum.
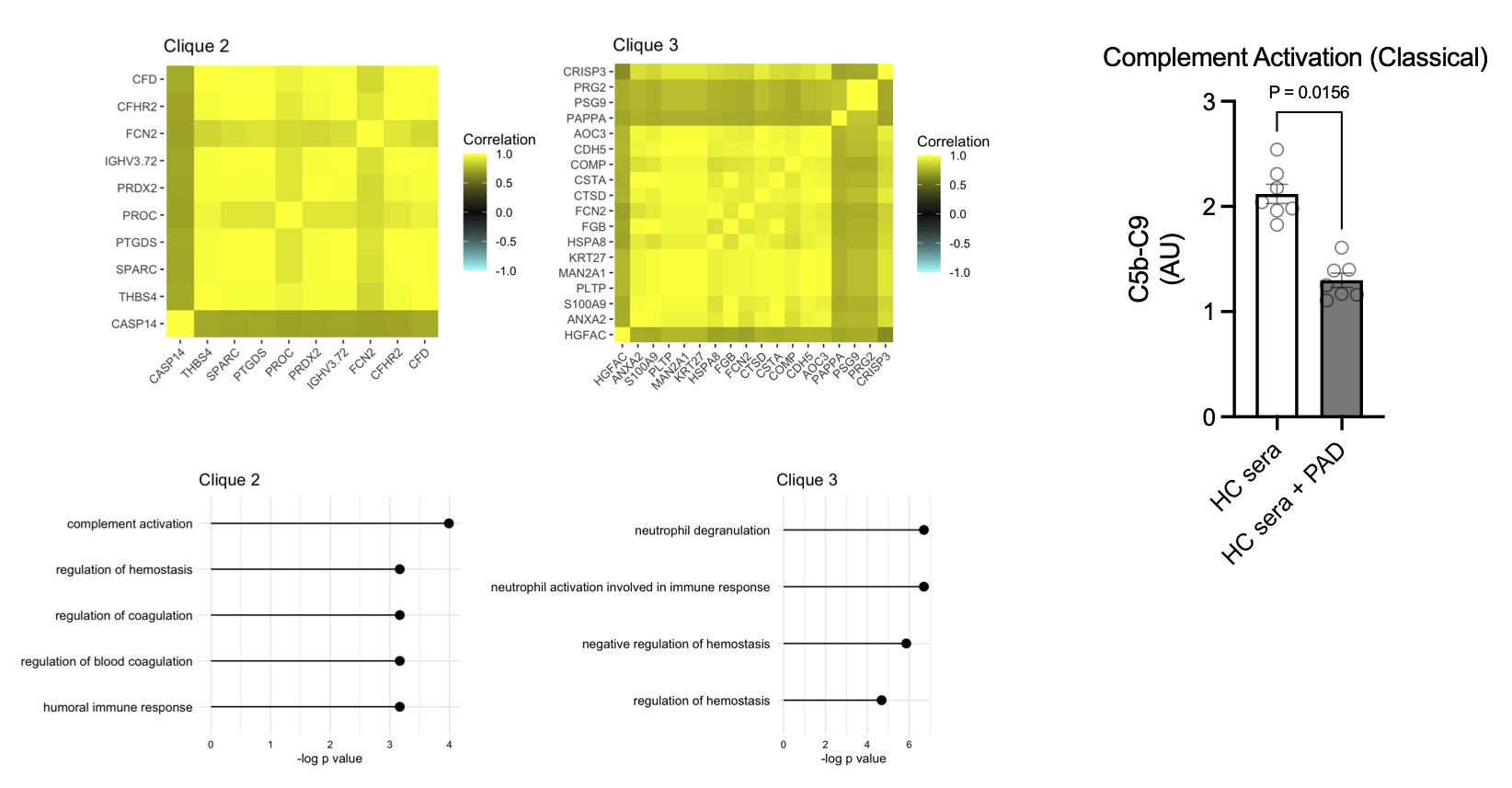
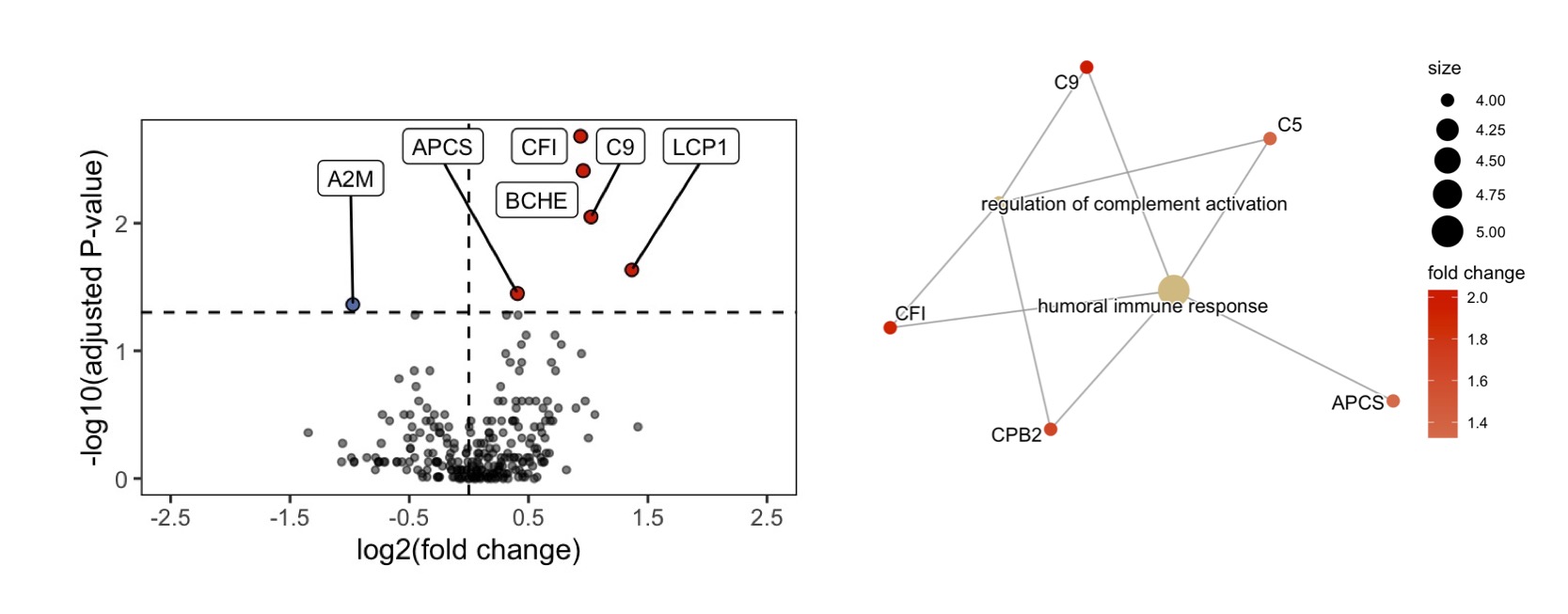
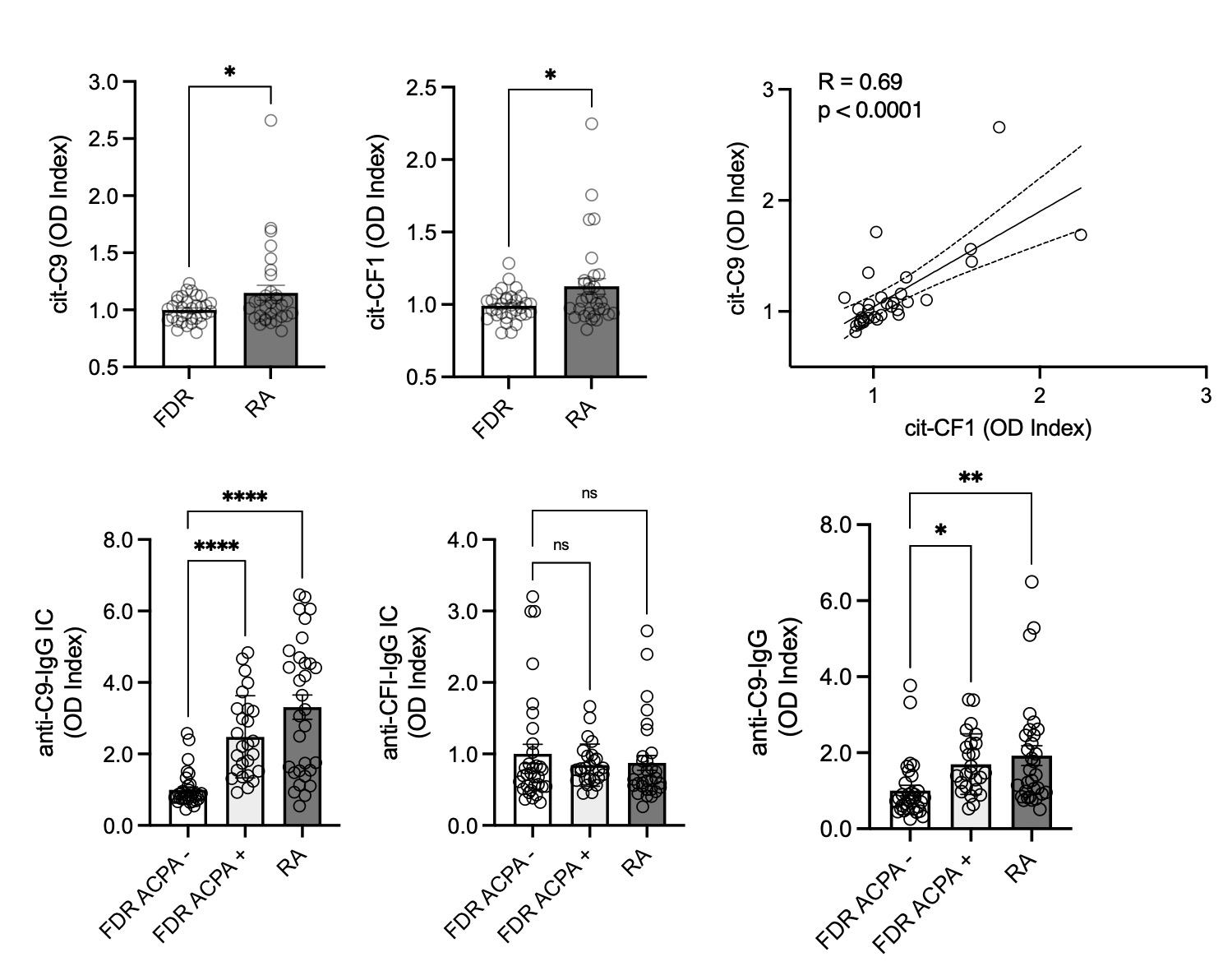
Results: After processing, a total of 249 cit-proteins were identified with high confidence. Using a machine learning algorithm called graph adjacency we identified 3 proteomic cliques (highly correlated protein sets, threshold R=0.7) in the human citrullinome. GO pathways were enriched in 2/3 cliques, with clique 2 enriched for complement activation and clique 3 enriched for neutrophil degranulation. Citrullination of HC sera (n=6) with PAD2 significantly reduced classical complement activation (p=0.016) compared to untreated HC sera (Fig1). The RA citrullinome displayed higher expression of complement proteins such as C9, CFB, CD1 and C5, and GO pathway enrichment included complement activation (Fig2). Total serum cit-C9 (p=0.02) and cit-CF1 (p=0.019) were validated with 63 samples (RA n=31) with a strong correlation (r=0.69, p< 0.0001) between cit-C9 and cit-CF1. Cit-C9 IgG ICs were elevated in ACPA+ FDR (n=26, p< 0.0001) and RA (p< 0.0001) compared to ACPA- FDR, but not cit-CF1. Autoantibodies to cit-C9 were also higher in ACPA+ FDR (p=0.01) and RA (Fig3, p=0.002).
Conclusion: This analysis of the circulating citrullinome in RA patients and ACPA+ at risk individuals suggests that augmented citrullination of specific proteins networks involved in complement activation and neutrophil degranulation may have direct functional consequences for these processes. The reduction in complement activation induced by PAD2 meditated citrullination may serve as a homeostatic mechanism to control the immune response, but also lead to the development of RA autoantibodies.
To cite this abstract in AMA style:
Trivedi K, Klenke C, Kim J, Maisha J, Sememenko A, MENG X, Navarrete M, El-Gabalawy H, O'Neil L. Proteomic Analysis of the Rheumatoid Arthritis Citrullinome Reveals an Enrichment of Citrullinated Complement Proteins [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/proteomic-analysis-of-the-rheumatoid-arthritis-citrullinome-reveals-an-enrichment-of-citrullinated-complement-proteins/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/proteomic-analysis-of-the-rheumatoid-arthritis-citrullinome-reveals-an-enrichment-of-citrullinated-complement-proteins/