Session Information
Date: Tuesday, November 14, 2023
Title: (2257–2325) SLE – Diagnosis, Manifestations, & Outcomes Poster III
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic lupus erythematosus (SLE) is the prototypical autoimmune disease with diverse clinical manifestations and variable treatment response. Conventional biomarkers of SLE activityprovide modest correlation with disease activity and suffer from several confounding factors that limit their utility as treat-to-target surrogates.The SLEDisease Activity Index 2000 (SLEDAI-2K) is often employed to monitor disease activity, but the administrative burden it requires has limited its real-world adoption.Modeling the values of serum biomarkers to predict patients’clinical SLEDAI-2Kscores (cSLEDAI-2K; which excludes its immunologic lab components, i.e., anti-dsDNA and complement), holds potential to advance the developmentof individualized treatment plans, thus advancing the field of precision medicine. Therefore, the aim of this study was to investigate the correlation of serum biomarkers with cSLEDAI-2K scores and to evaluate their potential to stratify patients in active and inactive disease states.
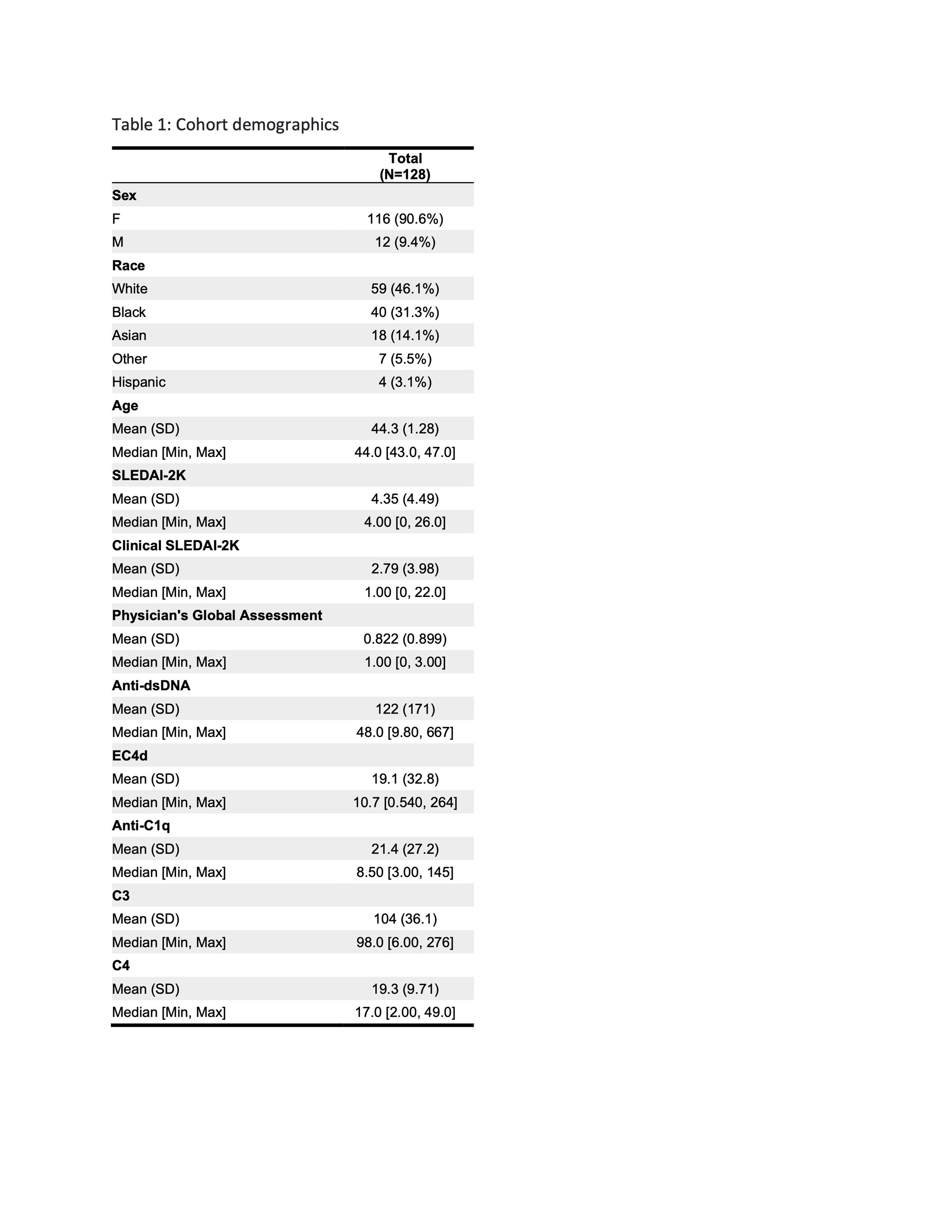
Methods: A total of 128 patients (90.6% female) with a mean age of 44.3 were included in the analysis. The patients sampled were raciallydiverse,as detailed in Table 1. The mean cSLEDAI-2K score was 4.35. The association of blood biomarkers, including anti-dsDNA, EC4d, PC4d, anti-C1q, C3, and C4with the cSLEDAI-2Kscore was evaluated using multiple 10-fold cross-validated regression models. The most predictive model was identified and used for further stratification of patients based on cSLEDAI-2Kscores of <4 (low activity) and >4 (high activity) groups.Significant differences in blood levels between patient groups were estimated using the Wilcoxon test.
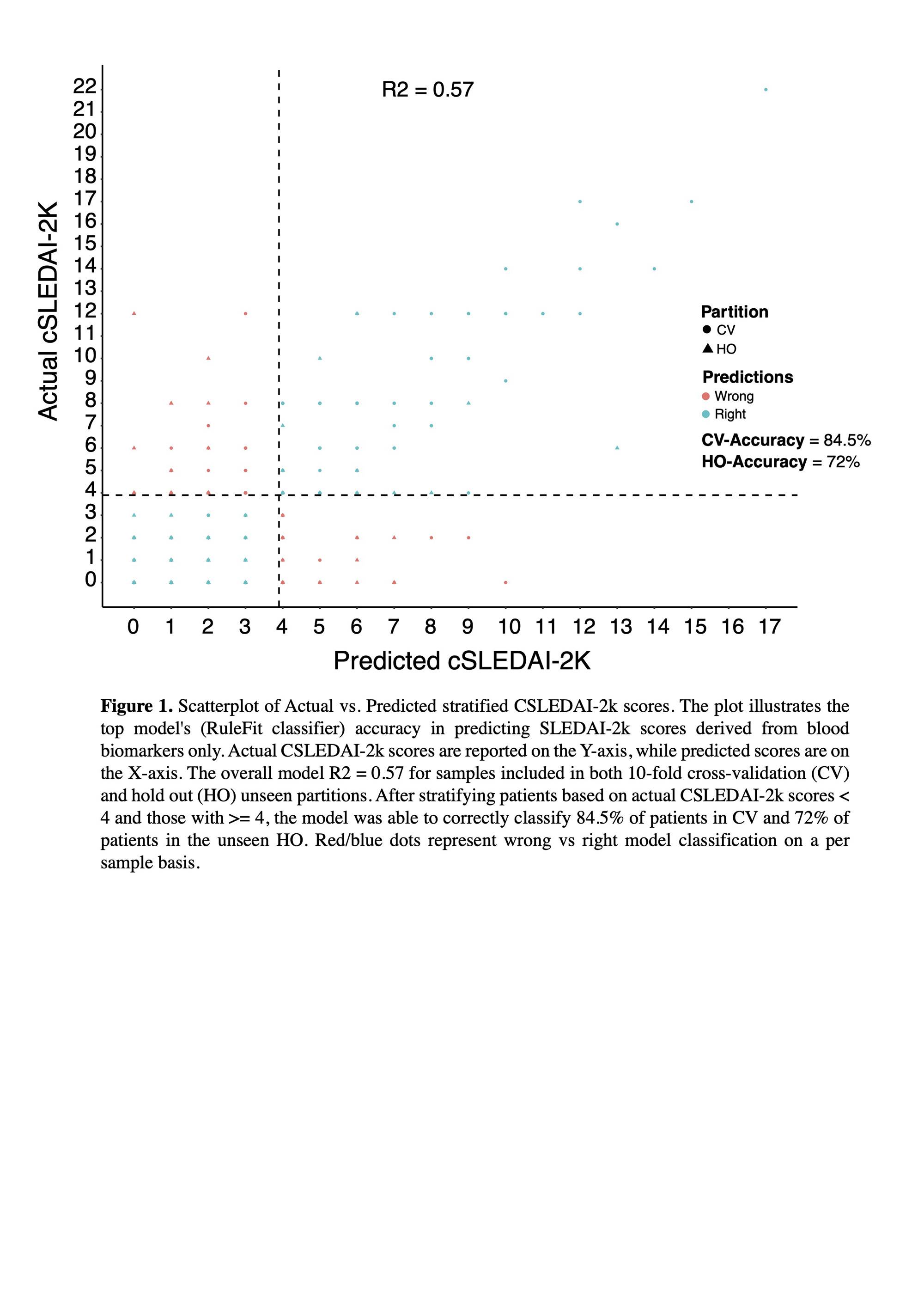
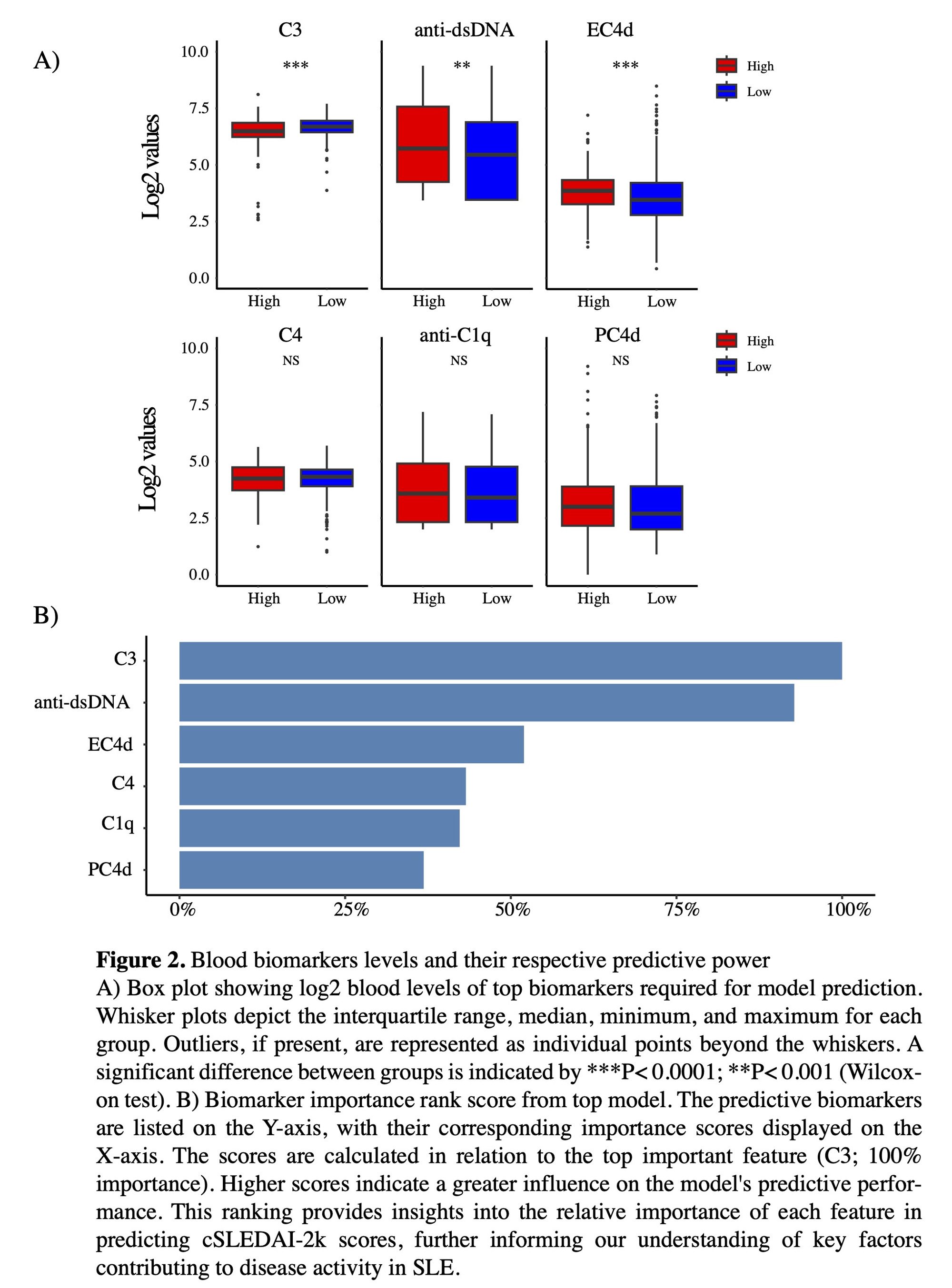
Results: The best-performing regression model (RuleFit Regressor) had an R2 of 0.57. Despite the modest correlation, the average absolute residual between actual and predicted cSLEDAI-2Kwas only 1.3. Moreover,when the predicted scores were used to categorize patients into cSLEDAI-2K <4 or >4, the model achieved an accuracy of 84.5% in cross-validation and 72% in the unseen holdout partition (Figure 1). Biomarkers were ranked in order of predictive importance showing C3 at the top, with 100% of relative model importance, followed by the other biomarkerspresentedin relation to C3 (Figure 2,B). Finally, comparing blood levels of the ranked biomarkers confirmed a statistically significant difference between SLE patients stratified bycSLEDAI-2K scores for C3 (P<0.001), anti-dsDNA (P= 0.005), and EC4d (P<0.001)(Figure 2, A), while differences inC4, anti-C1q, and PC4d were non-significant.
Conclusion: These results highlightthe potential of applying machine learning toselectedblood biomarkers to predict the cSLEDAI-2K score to monitor disease activity in clinical practice.EC4d emergedas anindependentpredictor,additive to the predictive power of conventional biomarkers across our top models, suggesting its potential use in a precision medicine approachto guide individualized treatment strategies in SLE. Further research is required to validate these findings and better understand their implications for individualized SLE management.
To cite this abstract in AMA style:
Kyttaris V, O'Malley T, Casaburi G, Kumar S, Concoff A. Predicting SLE Disease Activity with Blood Biomarkers: A Step Towards Precision Medicine [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/predicting-sle-disease-activity-with-blood-biomarkers-a-step-towards-precision-medicine/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/predicting-sle-disease-activity-with-blood-biomarkers-a-step-towards-precision-medicine/