Session Information
Date: Saturday, November 7, 2020
Title: RA – Diagnosis, Manifestations, & Outcomes Poster II: Biomarkers
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Hematoxylin and eosin (H&E) stained rheumatoid arthritis synovium are routinely used to assess inflammation [1]. In this work, we propose an automated approach using artificial intelligence (AI) to quantify all cell nuclei in whole slide H&E stained images. This approach provides quantitative information on the number and density of nuclei in synovial images. Given nuclei density is generally increased in sites of inflammation, this approach can identify areas of interest for further evaluation [2].
Methods: The proposed AI algorithm was used to segment cell nuclei in H&E-stained images. First, we split the whole slide image into 1024×1024 tiles with a resolution of 50 μm. Each tile was then processed as follows: 1) separate stained components using color deconvolution, 2) binarize image using local adaptive thresholding, 3) split clustered nuclei using the watershed algorithm, and 4) remove false-positive detected nuclei using shape analysis. Finally, the total nuclei count normalized for area of tissue and the nuclei density were calculated. In total, 166 H&E stained images were analyzed, and corresponding synovial tissue samples were classified by pathologist score of lymphocytic inflammation (0-4). Of which, 35 samples were also classified by RNA sequencing gene expression cluster (low, mixed, and high inflammatory gene expression) as previously described [1]. A one-way ANOVA test and post-hoc analysis using the Tukey’s test corrected for multiple comparisons were conducted.
Results: Upon visual inspection, the algorithm identified majority of the nuclei in a tile, as seen in Figure 1. There was a statistically significant difference in the mean nuclei count and nuclei density between the gene expression subtypes (p=0.01, p=0.03) as well as pathologist scores of lymphocytic infiltration (p< 0.01, p< 0.01), as seen in Table 1 and Figure 2. The post-hoc analysis revealed that the mean normalized nuclei count and nuclei density was significantly different between the mixed and high-inflammatory subtypes (p=0.01, p=0.03) and between the lowest and highest lymphocyte pathologist scores (p< 0.01, p< 0.01).
Conclusion: Automated image quantification of H&E stained nuclei suggests that nuclei count and density are increased in synovial samples with high inflammatory gene expression and high pathology scores of synovial lymphocytic inflammation. Additional algorithm validation is needed to better understand its limitations. This approach provides a quantitative framework to score stained images and identify abnormal or inflamed areas. Future efforts will attempt to identify other histological features which might be clinically useful for assessing RA synovium.
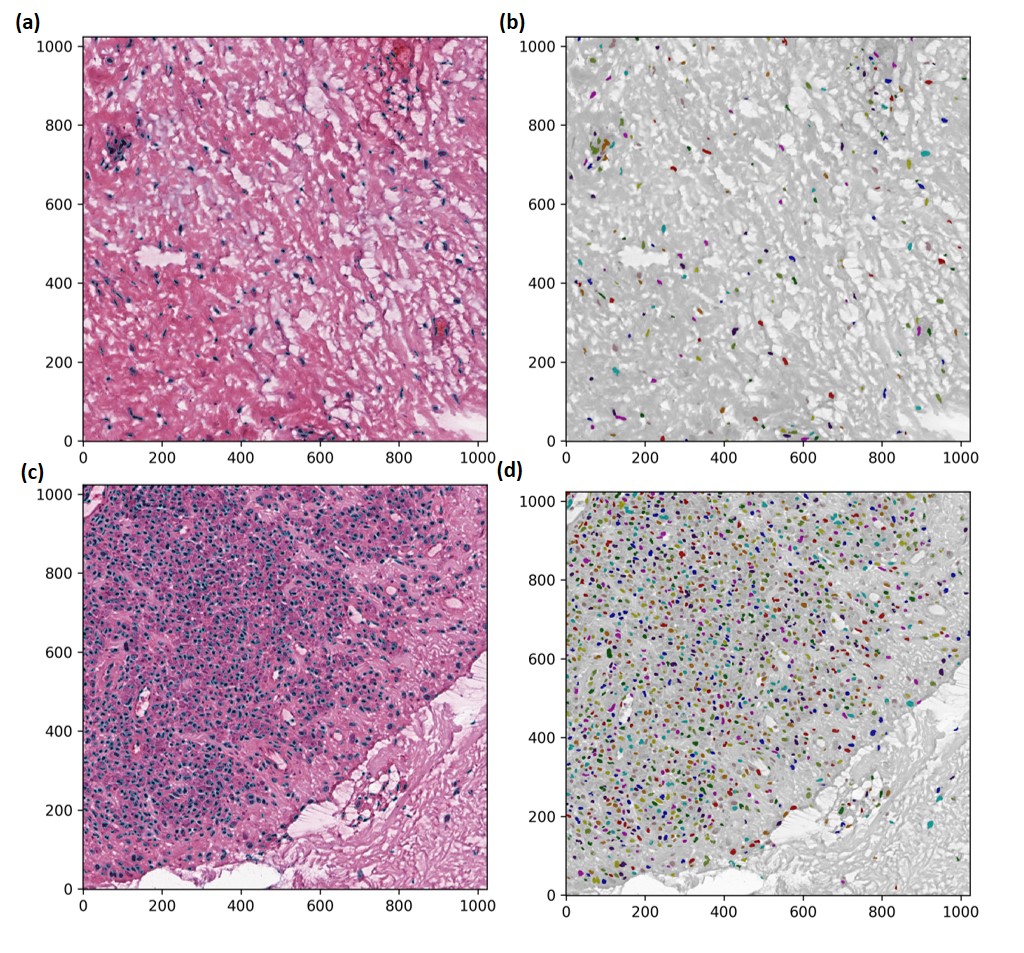 Figure 1. Nuclei identified using the proposed AI algorithm and visualized with boundaries and masks for tiles with mild (a-b) and band-like (c-d) lymphocytic inflammation.
Figure 1. Nuclei identified using the proposed AI algorithm and visualized with boundaries and masks for tiles with mild (a-b) and band-like (c-d) lymphocytic inflammation.
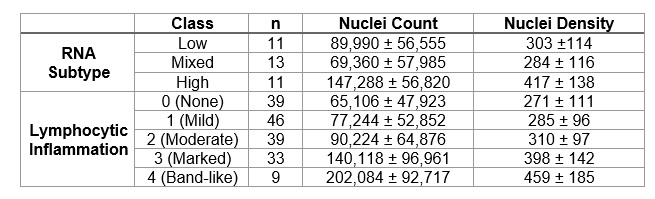 Table 1. RA cohort description of nuclei count and density by RNA subtypes and pathologist lymphocytic inflammation scores.
Table 1. RA cohort description of nuclei count and density by RNA subtypes and pathologist lymphocytic inflammation scores.
 Figure 2. Boxplots to visualize distribution of nuclei count and density by RNA subtypes (low, mixed, and high inflammatory) (a-b) and lymphocytic inflammation scored by pathologist (c-d).
Figure 2. Boxplots to visualize distribution of nuclei count and density by RNA subtypes (low, mixed, and high inflammatory) (a-b) and lymphocytic inflammation scored by pathologist (c-d).
To cite this abstract in AMA style:
Guan S, Slater D, Thompson J, DiCarlo E, Pearce-Fisher D, Goodman S, Mehta B, Orange D. Nuclei Detection in Rheumatoid Arthritis Synovial Tissue Using Artificial Intelligence [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/nuclei-detection-in-rheumatoid-arthritis-synovial-tissue-using-artificial-intelligence/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/nuclei-detection-in-rheumatoid-arthritis-synovial-tissue-using-artificial-intelligence/
