Session Information
Date: Sunday, November 7, 2021
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster (0541–0559)
Session Type: Poster Session B
Session Time: 8:30AM-10:30AM
Background/Purpose: While the etiology of systemic sclerosis (SSc) remains largely unknown, epigenetic dysregulation including abnormal DNA methylation is thought to contribute to its onset and progression. Currently, the most comprehensive assay for profiling DNA methylation is whole-genome bisulfite sequencing (WGBS), but its precision depends on read depth and it may be subject to sequencing errors. SOMNiBUS (Zhao et al. 2020) was developed to overcome some of these limitations. This one-stage method infers smooth covariate effects across regions while accounting for variable read depth, missing data patterns, sequencing errors, and confounders such as age. Using SOMNiBUS, we reanalyzed WGBS data on SSc patients and controls initially analyzed using bumphunter (Jaffe et al. 2012, Aryee et al. 2014, Lu et al., 2019), a less sensitive two-stage approach that fits single CpG associations first, followed by separate coefficient smoothing based on the fitted associations. We aimed to identify novel regions of dysregulated DNA methylation, thus providing a more comprehensive understanding of SSc pathogenesis.
Methods: Purified CD4+ T lymphocytes for 9 SSc and 4 control females were sequenced using WGBS. We separated the resulting sequence into regions with cuts made when adjacent CpG sites were spaced more than 200 bp apart. 8268 regions containing ≥ 60 CpGs were retained for analysis. DMRs were inferred with SOMNiBUS region-level test, adjusted for age. Pathway enrichment analysis was performed with Ingenuity Pathway Analysis (IPA) for genes with DMRs located on the gene body or within regulatory regions. We compared the results to those obtained by bumphunter in terms of plausibility of enriched pathways and agreement in genes detected.
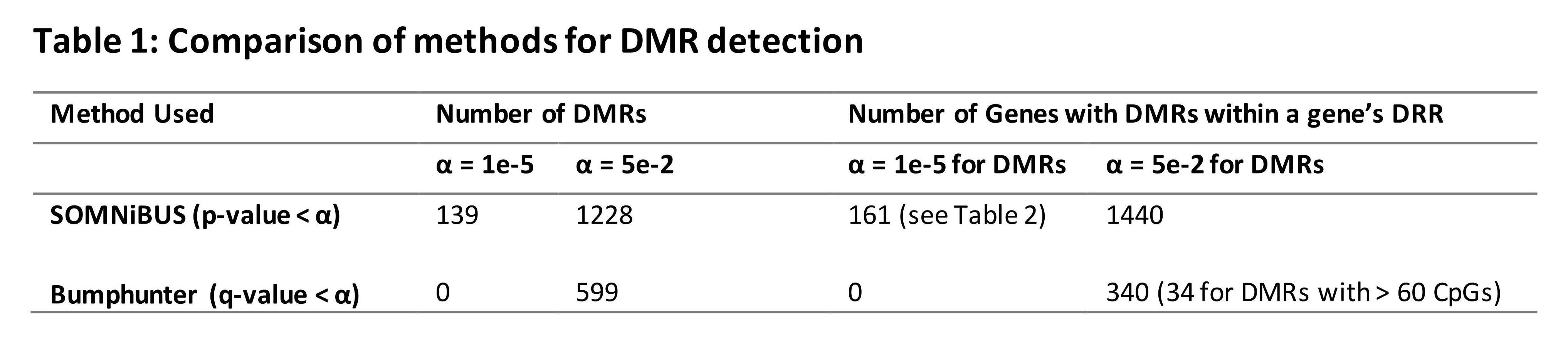
Results: We identified more DMRs with SOMNIBUS compared to bumphunter for both significance levels tested (Fig. 1 & Table 1). We selected all DMRs with a suggestive association defined by regional significance ≤ 1e-5 that overlapped either with a gene body or with the region 10,000 bp upstream of its transcription start site, allowing us to annotate DMRs up to the distal regulatory region (DRR), resulting in 161 differentially methylated genes (DMGs; Table 2). Changing the regional significance threshold to ≤ 0.05 for DMRs, we obtained 1,440 DMGs for SOMNiBUS compared to 340 DMGs for bumphunter (Table 1), 77 of which were identified by both methods. SOMNiBUS detected 29% of bumphunter’s DMGs with DMRs of < 60 CpGs, along with 22% of bumphunter’s other DMGs. In analyses stratified by autosomal and X chromosomes, the top-ranked autosomal gene was FLT4, a lymphangiogenic orchestrator previously associated with SSc (Manetti et al., 2019); 52 genes were identified on the X-chromosome (Table 2). IPA identified 10 enriched pathways for autosomal chromosomes with cell cycling as the top-ranked pathway and 5 enriched pathways for chromosomes X with antigen presentation as the top-ranked pathway (Table 2).
Conclusion: Using a novel and more powerful computational approach, we identified new and biologically plausible regions of the genome associated with SSc. These findings deepen biological insights into SSc and provide novel avenues of investigation into its pathogenesis.
To cite this abstract in AMA style:
Yu J, Lu T, Zhao K, Klein K, Lora M, Colmegna I, Greenwood C, Hudson M. Novel Insights into Systemic Sclerosis Using a Sensitive Computational Method to Analyze Whole-genome Bisulfite Sequencing Data [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/novel-insights-into-systemic-sclerosis-using-a-sensitive-computational-method-to-analyze-whole-genome-bisulfite-sequencing-data/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/novel-insights-into-systemic-sclerosis-using-a-sensitive-computational-method-to-analyze-whole-genome-bisulfite-sequencing-data/