Session Information
Date: Tuesday, October 23, 2018
Title: Systemic Lupus Erythematosus – Etiology and Pathogenesis Poster III
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose:
Neutrophil extracellular traps (NETs) are composed of decondensed DNA that is released during an active cell death program is known as NETosis. To date, florescence microscopy is the accepted method for quantification of NETs. However, this method is subjective, time consuming and yields low numbers of polymorphonuclear cells (PMNs) analyzed per sample. NETs contribute to pathogenesis of systemic lupus erythematosus (SLE) but the effects of disease activity on NETosis has not been adequately explored.
Methods:
Herein, we describe a flow cytometry-based assay for detection of NETosis within mixed cell populations and applied it to quantify NETting PMNs in SLE.
Results:
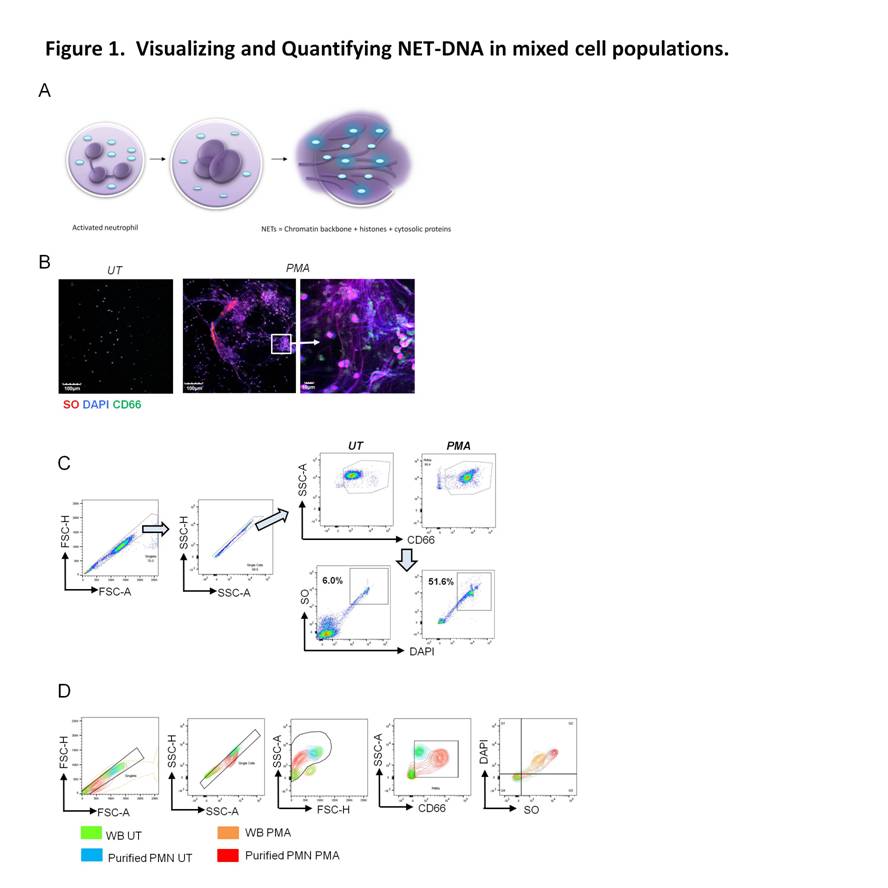
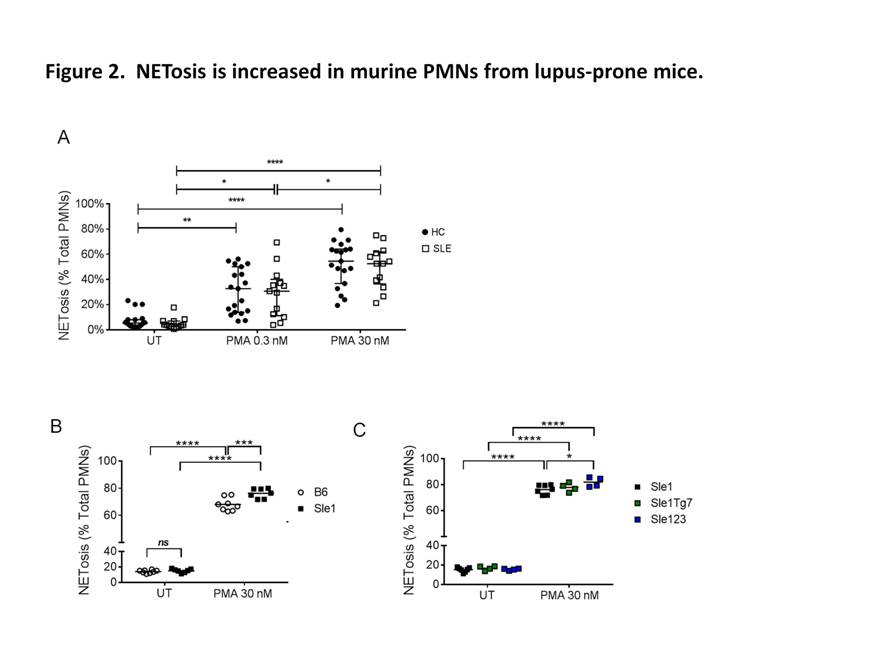
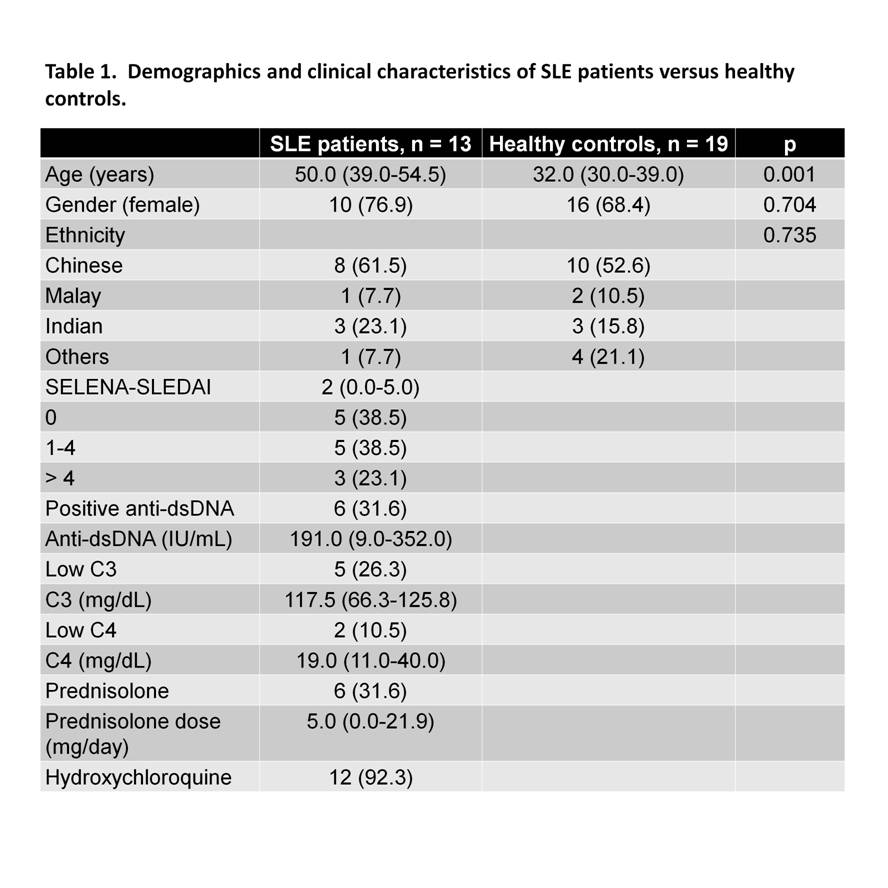
Using plasma membrane-impermeable DNA-binding dye, SYTOX Orange (SO), we found that cell-appendent DNA of NETting PMNs were positive for SO and DAPI (Fig. 1A-B). The combination of optimally diluted antibody and nucleic acid dyes required no washing and yielded low background fluorescence on flow cytometry (Fig. 1C-D). We then validated the assay by comparing with time-lapse live cell fluorescence microscopy and determined very good correlation (r = 0.90, P < 0.001), intra- and inter-assay variances, 3.98% and 4.66%, respectively. Significant correlations were found for NETosis from whole blood (WB) and purified PMNs (r = 0.79, P < 0.001). We examined PMA-induced NETosis in peripheral PMNs from SLE patients and controls and in bone marrow (BM) PMNs from multiple murine models (Fig 2A-C). However, most of the recruited SLE patients examined were on immunosuppressive therapy with quiescent disease activity (Table 1), which may have consequences for PMN activation (Fig. 2A). Therefore, we went on to examine NET formation in BM PMNs from young female lupus-prone mice. Stimulation with PMA resulted in an increase in the frequency of NETting BM PMNs, which was augmented in Sle1 mice, compared to B6 controls (Fig. 2B). An analysis of BM PMNs across all lupus-prone mice (Sle1, Sle1Tg7 and Sle123) showed an increase in NETosis in severe-lupus prone Sle123 mice compared to the milder lupus-prone Sle1 strain (Fig. 2C).
Conclusion:
This NETosis flow cytometry-based assay is observer-independent and allows for rapid assessment of a large number of PMNs in mixed cell populations. Disease activity may prime PMNs and lead to aberrant NETosis in SLE.
To cite this abstract in AMA style:
Tay SH, Zharkova O, Lee HY, Tripathi S, Ong WY, Lateef A, MacAry P, Lim L, Connolly JE, Fairhurst AM. Neutrophil Extracellular Trap Formation Is Dependent on Disease Activity in Systemic Lupus Erythematosus: Determination Using a Flow Cytometry-Based Assay [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/neutrophil-extracellular-trap-formation-is-dependent-on-disease-activity-in-systemic-lupus-erythematosus-determination-using-a-flow-cytometry-based-assay/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/neutrophil-extracellular-trap-formation-is-dependent-on-disease-activity-in-systemic-lupus-erythematosus-determination-using-a-flow-cytometry-based-assay/