Session Information
Session Type: ACR Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: Rheumatoid arthritis (RA) is the most prevalent autoimmune diseases (1-3% of the world’s population). RA is a prototypic inflammatory disease, being characterized by an altered state of homeostasis, in which immunological stimulation and unwanted inflammation prevail (Pincus, Brooks et al. 1994). The molecular triggers for the development of RA in healthy subjects are still not clearly defined. Identification of RA specific transcriptional signature can shed the light on the molecular pathogenesis of RA. Such signature facilitates the selection of biomarkers that can be used for early detection of RA and prediction of effectiveness of RA treatment. Most of the current transcriptome analyses in RA had focused on whole peripheral blood mononuclear cells (PBMCs). Few studies have indicated that natural killer (NK) cells may play either a pathogenic or a protective role in RA (Shegarfi, Naddafi et al. 2012, Yap, Tee et al. 2018). Hence, in this study we aimed at exploring NK cell markers that could possibly be used to aid in evaluating the differences among healthy controls and RA patients.
Methods: Publicly available transcriptome dataset (GSE93777) of sorted immune cells from RA patients and healthy volunteers were reanalyzed using in house pipeline for normalization and filtration of the raw probes’ expression. Gene Set Enrichment analysis was used to identify genes that are differentially expressed (DEG) between different immune cells as compared to NK cells. Next, DEG between NK cells of RA patients and healthy controls were selected. Both lists of identified DEG were validated in peripheral blood collected from healthy controls and RA patients. Whole blood samples were collected from the recruited 11 RA patients (satisfying the 2010 ACR/EULAR classification criteria for RA) and 10 healthy controls (with no diagnosis of autoimmune diseases). Then, PBMCs were separated by Ficoll density gradient method, while NK cells were isolated using RosetteSep negative selection method. RNA was extracted and gene expression was assessed using qRT-PCR. Statistical analysis was done using Mann-Whitney test. Binary regression test was used to predict the probabilities of being a case based on the values of the selected genes.
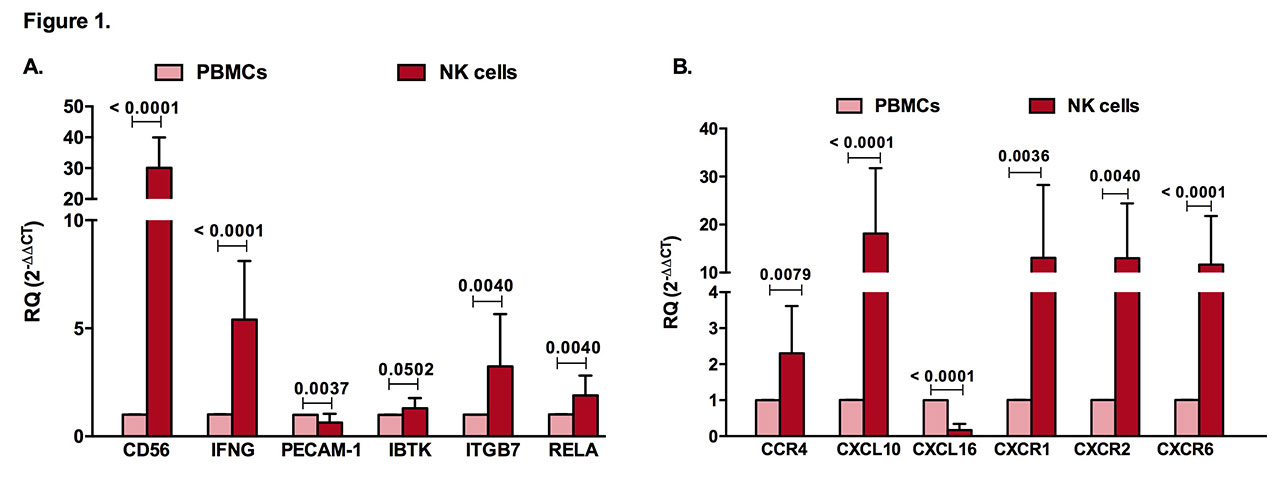
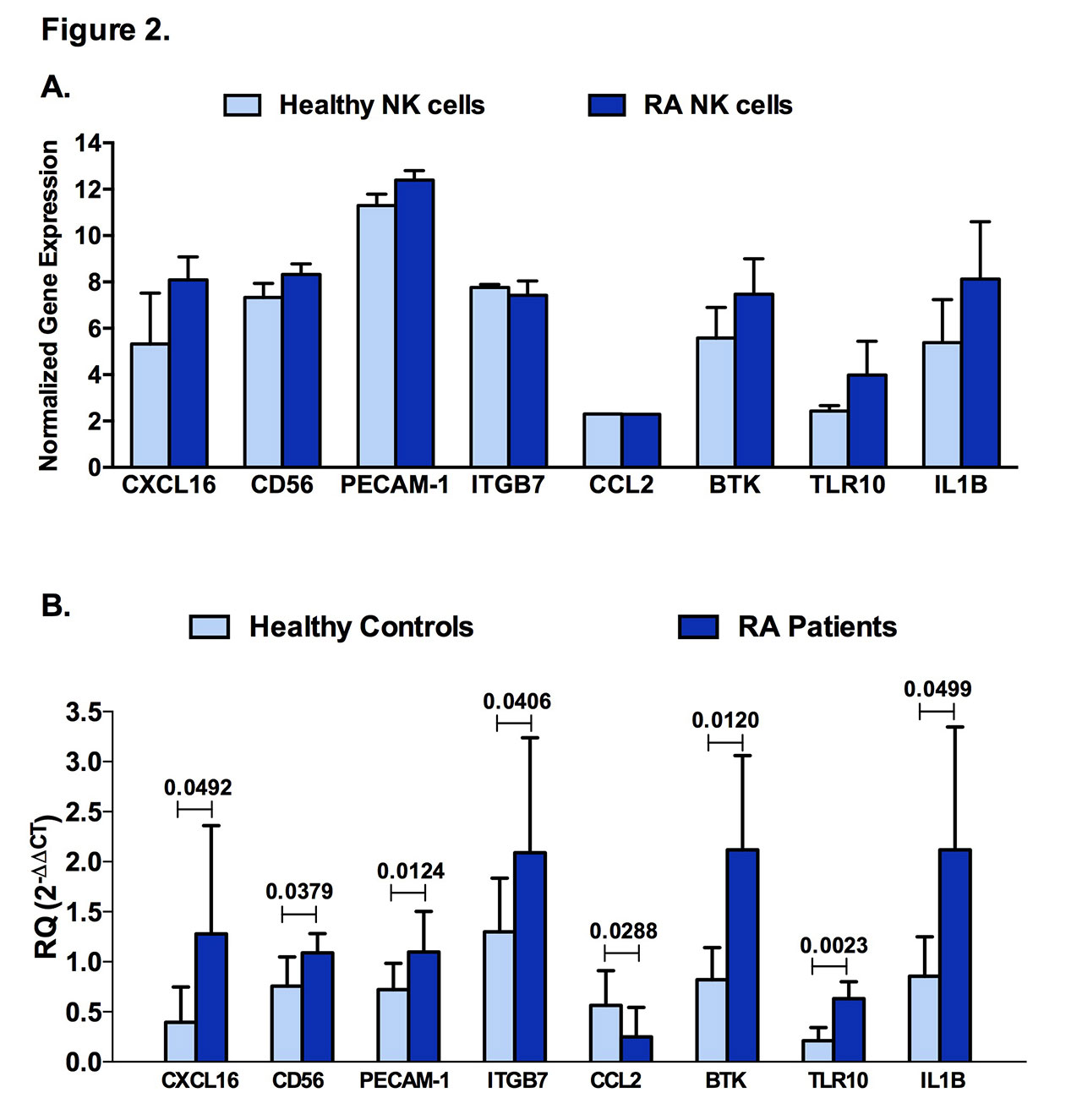
Results: Similar to CD56, a known marker for NK cells, and interferon gamma (IFN-γ), a prototype cytokine secreted by these cells, PECAM-1, IBTK, ITGB7, and RELA were expressed in NK cells (Figure 1A). Furthermore, chemokine ligands and receptors (CCR4, CXCL10, CXCL16, CXCR1, CXCR2, CXCR6) were specifically expressed by NK cells in comparison to PBMCs (Figure 1B). Additionally, as predicted by bioinformatics (Figure 2A), CXCL16, CD56, PECAM-1, ITGB7, BTK, TLR10, and IL-1β were significantly upregulated in NK cells of RA patients when compared to healthy controls, whereas CCL2 was downregulated (Figure 2B). In this study, we applied a logistic regression model using some of the identified NK genes expression (RELA, BTK, IL-1β, CKLF, PECAM-1, TLR10, IL1RN, IFN-γ) where it showed a good prediction power as predictors for RA.
Conclusion: Gene expression of these molecules; namely CXCL16, CD56, PECAM-1, ITGB7, BTK, TLR10 and IL-1β, in NK cells might be used as promising biomarkers for RA disease.
To cite this abstract in AMA style:
Elemam N, Hachim M, Hannawi S, Maghazachi A. Natural Killer Cells Gene Expression Can Differentiate Rheumatoid Arthritis Patients from Healthy Controls [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/natural-killer-cells-gene-expression-can-differentiate-rheumatoid-arthritis-patients-from-healthy-controls/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/natural-killer-cells-gene-expression-can-differentiate-rheumatoid-arthritis-patients-from-healthy-controls/