Session Information
Date: Monday, November 8, 2021
Title: Abstracts: Systemic Sclerosis & Related Disorders – Basic Science (1434–1437)
Session Type: Abstract Session
Session Time: 10:30AM-10:45AM
Background/Purpose: Discovery of biomarkers in systemic sclerosis (SSc) represents an unmet clinical need. Samples from prominently affected fibrotic end-organs such as lung and skin are not readily available whereas serum proteins are assessed during routine clinical care. However, it is unclear to what extent serum proteins reflect the molecular dysregulations of peripheral blood cells (PBCs) or affected end-organs. This study describes multiomic comparative analysis of SSc serum proteomic profile as well as gene expression analysis of PBC and skin biopsies in concurrently collected samples from SSc patients and healthy controls.
Methods: Global gene expression profiling on Illumina HumanHT-12 BeadChip was carried out in skin and PBC samples obtained from 49 patients and 25 unaffected controls. Levels of 911 proteins were determined in concurrently collected serum samples with Olink Proximity Extension Assay (PEA) technology.
Results: SSc serum profile revealed an upregulation of proteins involved in pro-fibrotic, homing and extravasation, as well as extracellular matrix components/modulators. Notably, several soluble receptor proteins such as EGFR, ERBB2, ERBB3, VEGFR2, TGFBR3, and PDGF-Rα were downregulated. Thirty-nine proteins correlated with severity of SSc skin disease as assessed by modified Rodnan Skin Score (mRSS) (FDR< 5%). There was a significant correlation between direction of differential expression of serum proteins in SSc vs. control comparison and their association with mRSS (ρ=0.59, permutation p< 0.0001) (Fig. 1). Correlative analysis of differences between SSc patients and healthy controls among the three datasets demonstrated that:
1) The differential expression of serum protein in SSc vs. control comparison significantly correlated with the differential expression of corresponding transcripts in skin but not in PBCs (p=0.014 vs. p=0.24) (Fig 2A).
2) The differentially expressed serum proteins represented more significant Well-Associated-Proteins (WAP) (Pradines et al. PMC7046299) in the skin than in PBC gene expression dataset, as demonstrated by lower WAP scores in the skin tissue (Fig. 2B).
3) The assessment of the concordance of between-sample similarities at the entire dataset level revealed that the molecular profile of serum proteins and skin gene expression data were significantly concordant in SSc patients but not in healthy controls. On the contrary, the concordance of similarities between serum protein and PBC gene expression profiles was most pronounced in healthy controls and notably reduced in patients, suggesting disruption of correlation between the serum proteome and PBC transcriptome in SSc patients, contrary to healthy controls due to spill-over effect from skin. (Fig 3).
Conclusion: In this first multiomic cross-comparison of SSc serum proteins, PBC transcriptome and skin transcriptome in concurrently collected samples, the serum protein profile correlated more closely with molecular dysregulations of skin than PBCs, supporting the notion that it might serve as a reflection of disease severity at the end-organ level.
 Serum proteins associations with disease and mRSS: a) Volcano plot of SSc-Cont differences; b) Volcano plot of correlation with mRSS in SSc patients; c) Scatterplot of serum proteins associations with mRSS vs. SSc-Cont differences; d) Permutation-based significance of the correlation between serum protein associations with mRSS and SSc-Cont differences. Vertical red dashes in panel d) indicate observed correlation between mRSS and disease effects on serum proteome; histogram represents distribution of such correlations obtained for randomly permuted sample annotation. Text labels in panel c) indicate proteins associated at BH-FDR < 5% both with mRSS and disease.
Serum proteins associations with disease and mRSS: a) Volcano plot of SSc-Cont differences; b) Volcano plot of correlation with mRSS in SSc patients; c) Scatterplot of serum proteins associations with mRSS vs. SSc-Cont differences; d) Permutation-based significance of the correlation between serum protein associations with mRSS and SSc-Cont differences. Vertical red dashes in panel d) indicate observed correlation between mRSS and disease effects on serum proteome; histogram represents distribution of such correlations obtained for randomly permuted sample annotation. Text labels in panel c) indicate proteins associated at BH-FDR < 5% both with mRSS and disease.
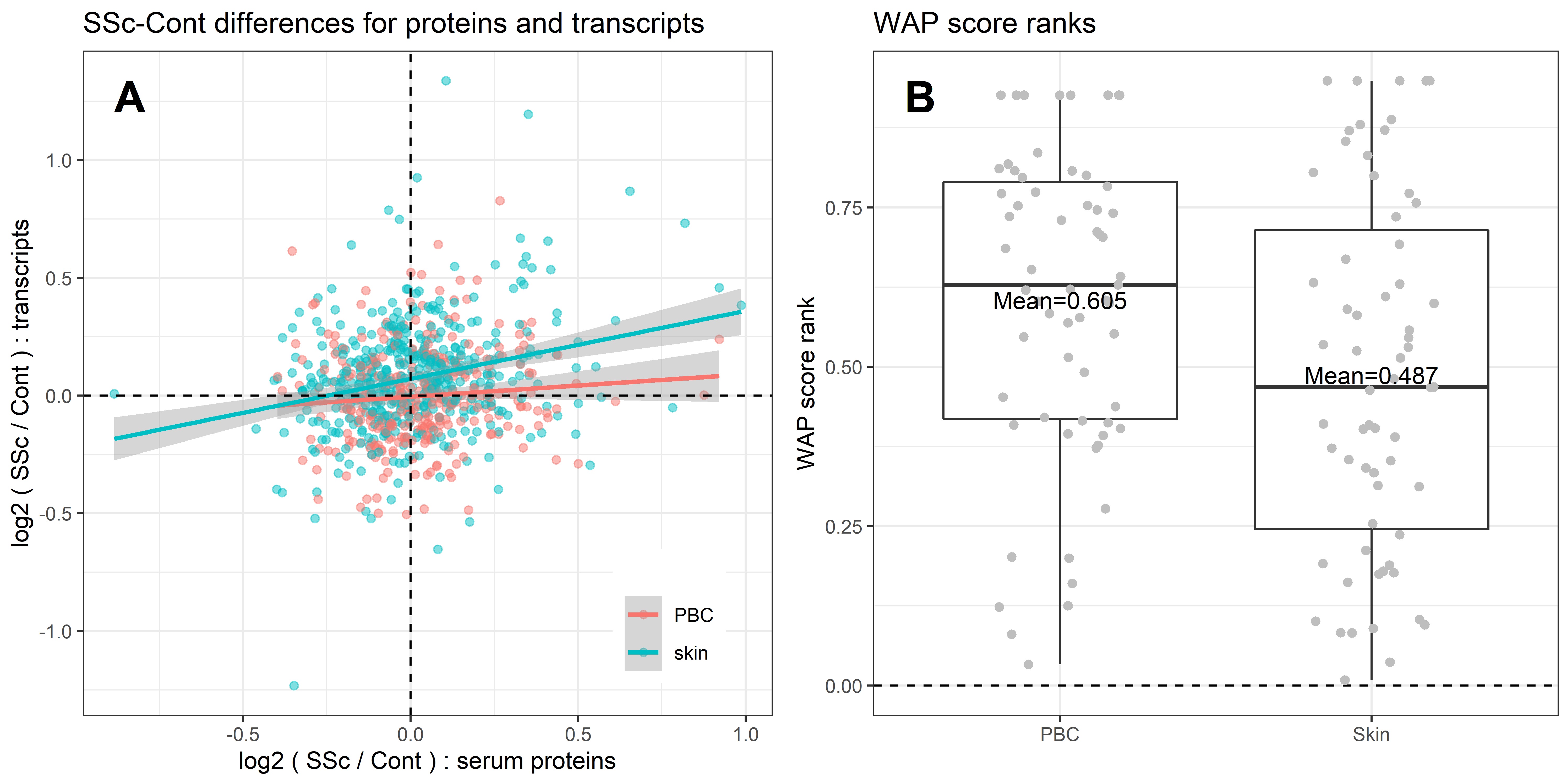 SSc-Cont differences for the serum proteins are significantly associated with SSc-Cont differences in skin: a) SSc-Cont differences are positively correlated between serum proteins and corresponding skin transcripts; b) WAP score ranks of differentially expressed serum proteins for SSc-Cont differences in skin and in PBC (lower values of rank represent more significant WAPs). The WAP ranking is generated based on prior knowledge in the form of protein-protein interaction networks.
SSc-Cont differences for the serum proteins are significantly associated with SSc-Cont differences in skin: a) SSc-Cont differences are positively correlated between serum proteins and corresponding skin transcripts; b) WAP score ranks of differentially expressed serum proteins for SSc-Cont differences in skin and in PBC (lower values of rank represent more significant WAPs). The WAP ranking is generated based on prior knowledge in the form of protein-protein interaction networks.
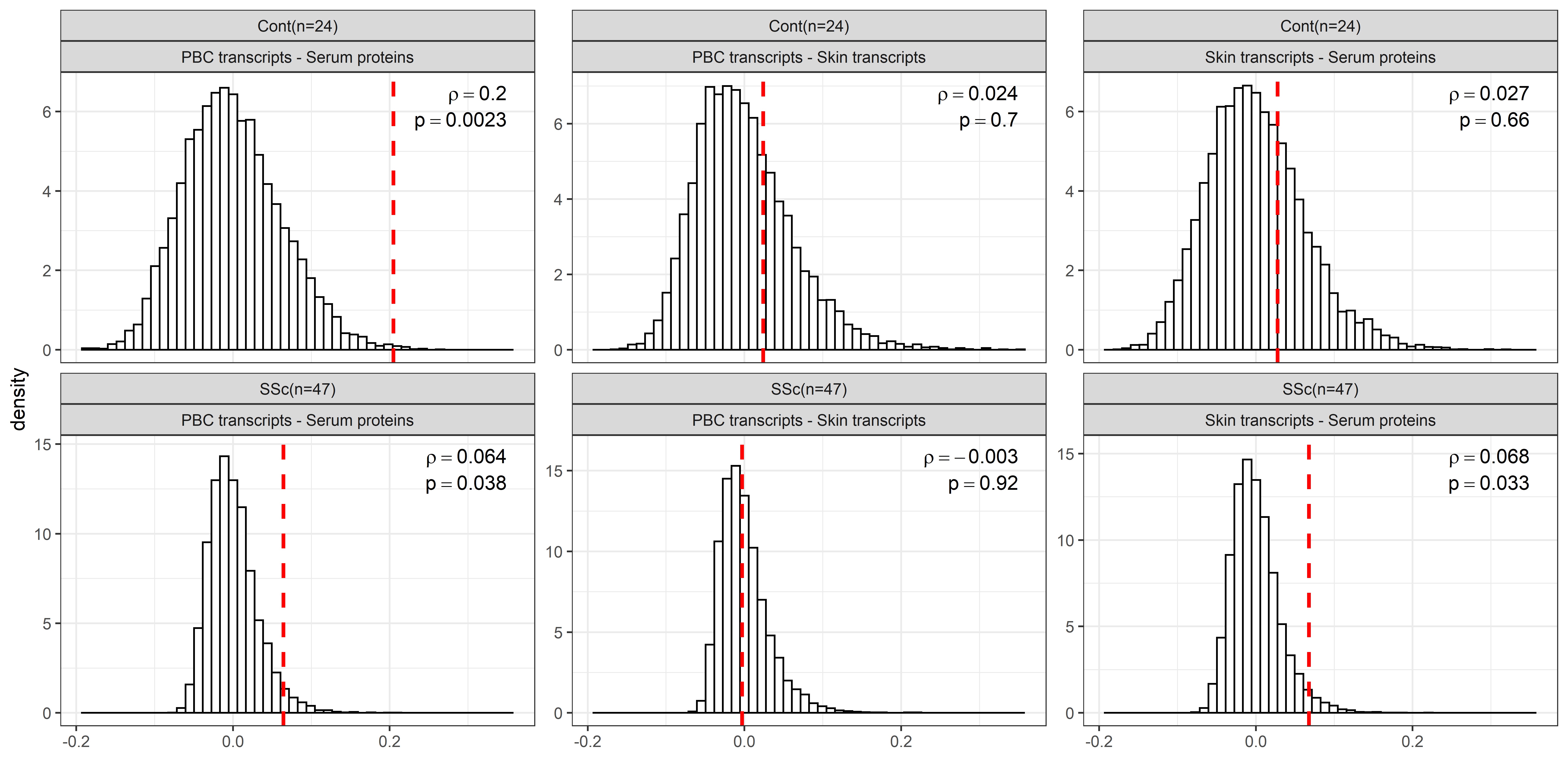 Concordance of between samples similarities (as Spearman correlations) for each pairwise combination of the three datasets: serum proteins, PBC transcripts and skin transcripts. Top row displays results for healthy controls, bottom row – for SSc patients. Mantel test results (vertical red dashes represent the observed concordance of between sample similarities for the actual mapping of samples to subjects) are compared to their corresponding null distributions (obtained by randomly permuting assignment of samples to subjects in each dataset).
Concordance of between samples similarities (as Spearman correlations) for each pairwise combination of the three datasets: serum proteins, PBC transcripts and skin transcripts. Top row displays results for healthy controls, bottom row – for SSc patients. Mantel test results (vertical red dashes represent the observed concordance of between sample similarities for the actual mapping of samples to subjects) are compared to their corresponding null distributions (obtained by randomly permuting assignment of samples to subjects in each dataset).
To cite this abstract in AMA style:
Farutin V, Kurtagic E, Pradines J, Capila I, Mayes M, Wu M, Manning A, Assassi S. Multiomic Study of Skin, Peripheral Blood and Serum: Is Serum Proteome a Reflection of Disease Process at the End-Organ Level in Systemic Sclerosis? [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/multiomic-study-of-skin-peripheral-blood-and-serum-is-serum-proteome-a-reflection-of-disease-process-at-the-end-organ-level-in-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/multiomic-study-of-skin-peripheral-blood-and-serum-is-serum-proteome-a-reflection-of-disease-process-at-the-end-organ-level-in-systemic-sclerosis/
