Session Information
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Abatacept (ABA) is a fusion protein that disrupts T cell co-stimulation by inhibiting interactions between CD28 and CD80/86. ABA is effective in RA, but SLE clinical trials have been disappointing. Exploratory analyses have suggested that ABA might benefit some SLE patients depending on disease severity or outcome definitions, but identification of reproducible predictors of response remains elusive. This study applied a systems-based approach to developing a predictive model of abatacept response in SLE.
Methods: Pre-treatment peripheral blood mononuclear cells were obtained from the screening visits of the Clarification of Abatacept Effects in SLE with Integrated Biologic and Clinical Approaches (ABC; n=19) and the Abatacept and Cyclophosphamide Combination Therapy for Lupus Nephritis (ACCESS; n=40) clinical trials evaluating ABA in SLE patients without organ-threatening disease or lupus nephritis, respectively. Genomic DNA and total RNA were extracted from isolated T cells, B cells, and monocytes, followed by DNA methylation microarray and RNA sequencing analyses. Machine learning was applied to methylation data using generalized linear modeling and 40 bootstrapped re-samplings split between 70% training and 30% testing.
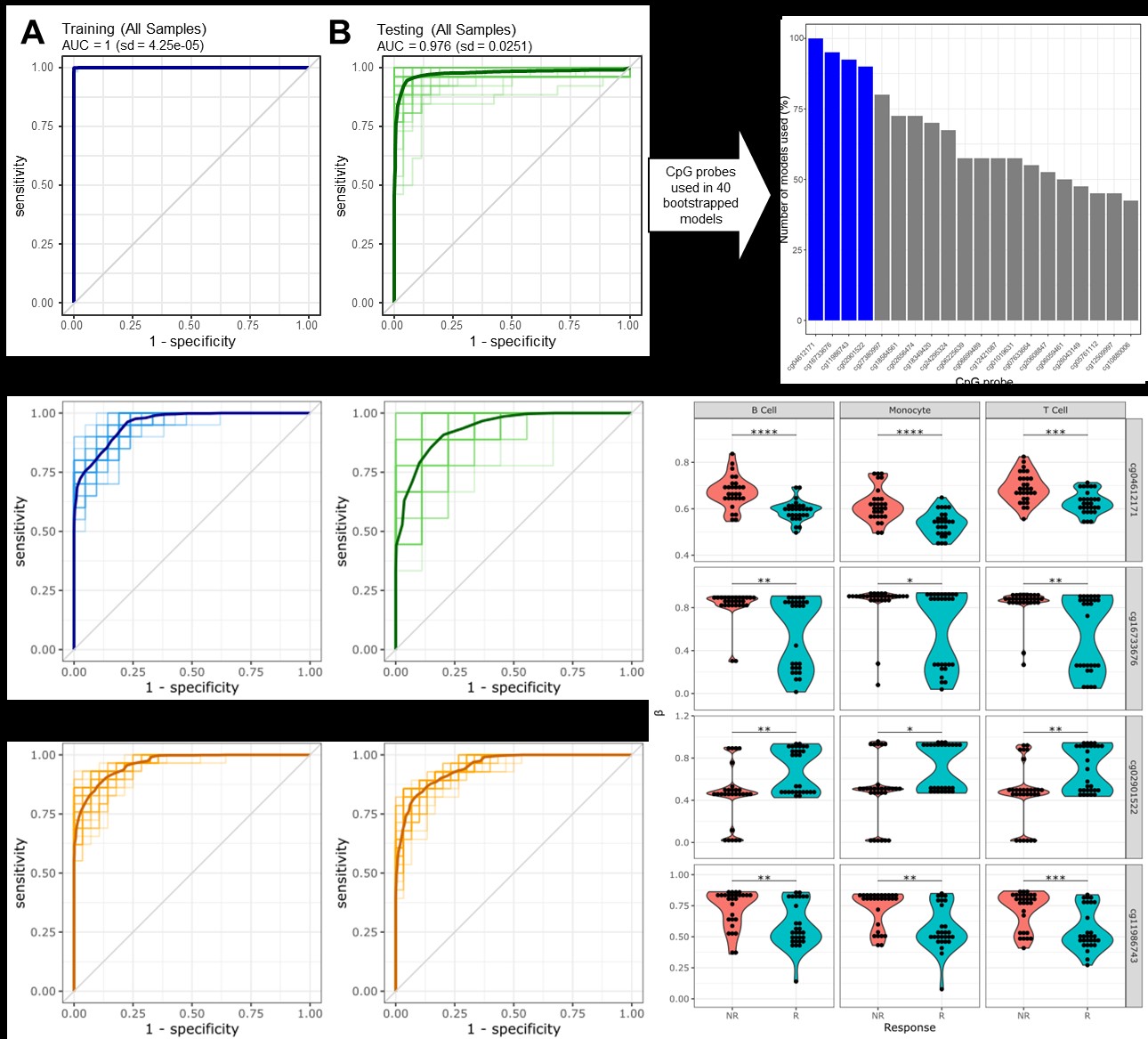
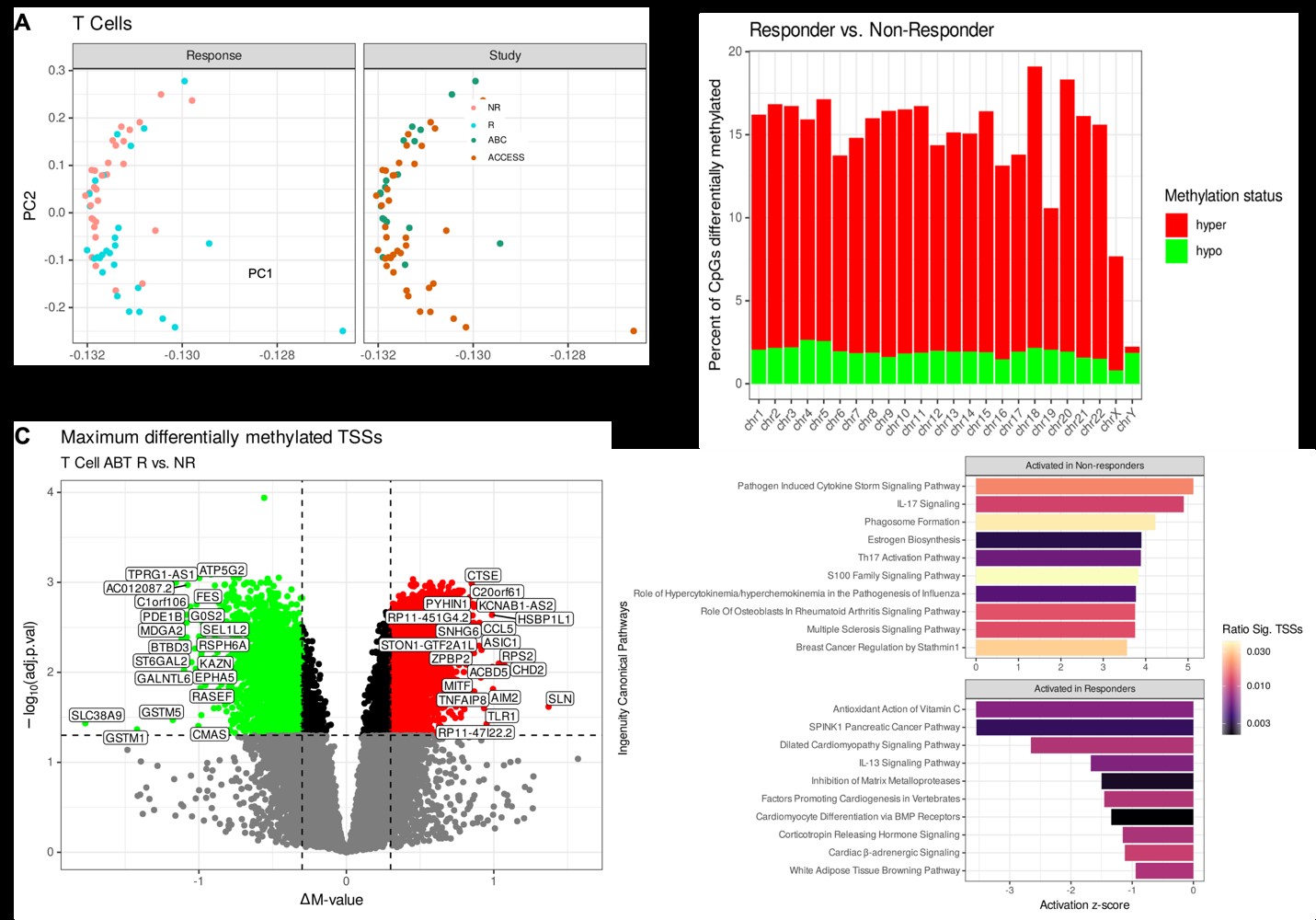
Results: A total of 29 ABA responders (R) and 30 non-responders (NR) were identified from both clinical trials. Bootstrapped models of all cell types separated responders and non-responders robustly, with an average AUC of 0.976 (Fig 1A-B). Four CpGs were shared in at least 80% of the re-samplings (Fig 1C), and a model using these 4 CpGs on T cells alone revealed an average ROC of 0.935 (Fig. 1D-E). Applying this model to B cells and monocytes yielded similarly robust results (Fig. 1F-G), and the 4 CpGs were differentially methylated in each cell type (Fig. 1H). In T cells, the 4 CpGs positively correlated with TCR activation, CTLA-4 regulation, and IL-2 production (Fig 2A-B), while BCR activity and phagosome activity and cytokine production are correlated with these CpGs in B cells and monocytes, respectively (Fig 2C-F). Unbiased principal component analysis on global T cell methylation profiles separated R and NR patients (Fig 3A), and most sites were hypermethylated in R patients (Fig 3C). The most hypermethylated transcription start sites revealed associations with cytokine storm and IL-17 signaling in NR patients (Fig 3D-E).
Conclusion: We determined a robust 4 CpG methylation signature that predicts ABA response across two disparate SLE trials. Parallel transcriptomic profiling linked this signature to cell lineage-specific activation pathways. Comparative epigenomic analysis of T cells revealed a hypomethylated signature in NR patients, likely driving continued proinflammatory responses despite abatacept treatment.
To cite this abstract in AMA style:
Thomas K, Smith M, Guthridge C, Dominguez N, Macwana S, DeJager W, Wagner C, Schafer P, Kamp S, Diamond B, Wofsy D, Thanou A, Arriens C, Aranow C, Merrill J, James J, Guthridge J. Molecular Predictors of Treatment Response in Two Trials of Abatacept in Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/molecular-predictors-of-treatment-response-in-two-trials-of-abatacept-in-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/molecular-predictors-of-treatment-response-in-two-trials-of-abatacept-in-systemic-lupus-erythematosus/