Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Autoimmune and immune-mediated diseases (AIMDs) affect over 20 million Americans. The sharp increase in prevalence of these disorders over recent decades suggests that factors other than genetics may be involved in pathogenesis. While the microbiome has been previously investigated as a risk factor, most studies have considered AIMDs in isolation. Here, we hypothesize that analysis across patients with different AIMDs will identify protective and deleterious microbial factors that are unique or shared across diseases.
Methods: As a part of the Accelerating Medicines Partnership-Autoimmune and Immune Mediated Diseases (AMP AIM) consortium, we characterized the microbiome of 126 subjects across six AIMDs: rheumatoid arthritis (RA; n=43), psoriasis (PsO=28), psoriatic arthritis (PsA=28), Sjögren’s disease (SjD=10), non-Sjögren’s sicca (NSS=12), and systemic lupus erythematosus (SLE=5), as well as 6 healthy controls. Four previously published studies in which data was publicly available were used as validation cohorts (Table 1). All studies consisted of 16S rRNA sequencing and were analyzed using QIIME2, the GreenGenes2 taxonomy, and LEfSe. Statistical tests (Wilcoxon Rank-Sum, Kruskal-Wallis) were performed in R 4.4.2 using the base stats library, and comparisons of features across groups was visualized using UpSetR.
Results: Within the AMP AIM cohort, AIMD patients exhibited significantly lower alpha diversity (p< 0.05) and significant differences in beta diversity compared to healthy controls for all diseases (except for SLE where p=0.08) (Figure 1). LEfSe identified taxa that were differential in disease conditions; Lawsonibacter was enriched in SjD patients, while Anaerotignaceae and Othenecus were depleted in PsO, RA, and SLE patients. Across all six AIMDs, Phascolarctobacterium was significantly depleted relative to healthy controls. In four validation datasets, when comparing disease groups to healthy controls, we observed significantly reduced alpha diversities in one study (Valid2 SjD) and significantly different beta diversities in all four. Lawsonibacter was enriched in the affected patients within the Valid1 SLE, Valid2 SjD, and Valid3 RA studies. Conversely, Anaerotignaceae and Othenecus, were depleted in the Valid1 SLE and Valid2 SjD studies. Additionally, Phascolarctobacterium exhibited lower mean abundances in the disease subgroups relative to healthy controls in Valid1 SLE and Valid3 RA studies (although not significantly).
Conclusion: In our AMP AIM cohort and across validation studies, comparison of healthy controls and patients with AIMDs revealed differences in alpha diversity, beta diversity, or both. We identified several taxa commonly enriched (Lawsonibacter) and depleted (Phasolarctobacterium, Anaerotignaceae, and Othenecus) across multiple AIMDs in both AMP AIM and validation studies. Across studies, there was a greater overlap among taxa that were depleted across disease states than among taxa enriched across AIMDs. Our meta-analysis suggests that microbes associated with pathogenesis are likely unique to each disease, potential mechanisms of protection (or lack thereof) tend to be shared.
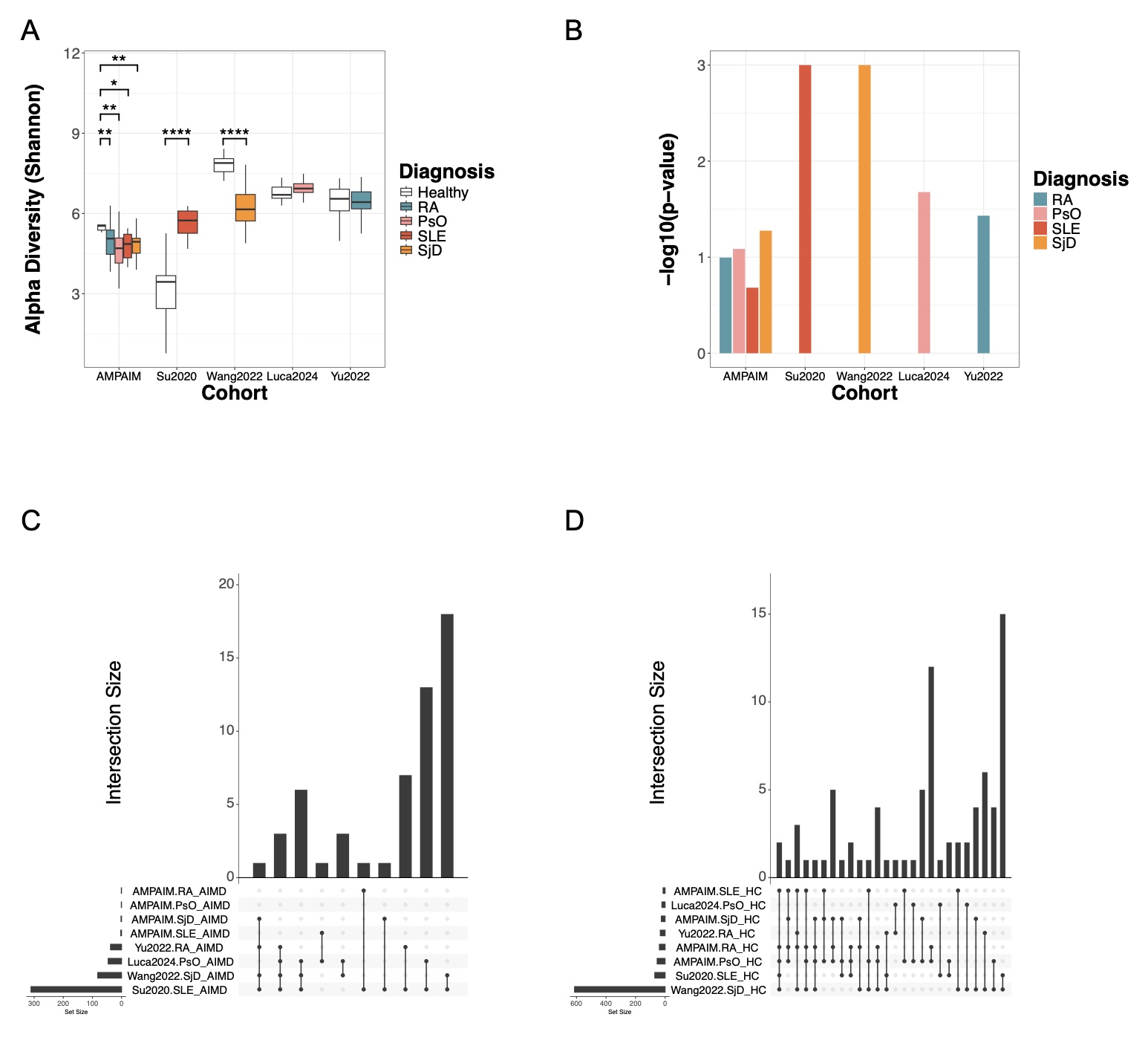 Table 1. Cohort sizes and breakdown of diagnoses in meta-analysis datasets.
Table 1. Cohort sizes and breakdown of diagnoses in meta-analysis datasets.
.jpg) Figure 1. Patients with AIMDs exhibit distinct microbiome compositions from healthy controls. (A) Alpha diversity (Shannon index) within the AMP-AIM cohort. (B) PCoA of beta diversity (Unweighted UniFrac) of AMP AIM cohort samples, *p < 0.05, **p < 0.01.
Figure 1. Patients with AIMDs exhibit distinct microbiome compositions from healthy controls. (A) Alpha diversity (Shannon index) within the AMP-AIM cohort. (B) PCoA of beta diversity (Unweighted UniFrac) of AMP AIM cohort samples, *p < 0.05, **p < 0.01.
.jpg) Figure 2. Healthy controls and patients with AIMDs exhibit distinct microbiome compositions across validation studies with overlap in differential taxonomic features. (A) Alpha diversity (Shannon index) of AMP AIM cohort along with 4 validation datasets, broken down by diagnoses. (B) PERMANOVA p-values (negative log-transformed) testing the difference in composition between AIMD and healthy control subgroups within each study for each diagnosis. (C-D) UpSetR plots showing the sizes of intersections of taxa identified as enriched (C) or depleted (D) in disease state relative to healthy controls across AMP AIM and validation cohorts.
Figure 2. Healthy controls and patients with AIMDs exhibit distinct microbiome compositions across validation studies with overlap in differential taxonomic features. (A) Alpha diversity (Shannon index) of AMP AIM cohort along with 4 validation datasets, broken down by diagnoses. (B) PERMANOVA p-values (negative log-transformed) testing the difference in composition between AIMD and healthy control subgroups within each study for each diagnosis. (C-D) UpSetR plots showing the sizes of intersections of taxa identified as enriched (C) or depleted (D) in disease state relative to healthy controls across AMP AIM and validation cohorts.
To cite this abstract in AMA style:
Bu K, Blank R, Boix-Amoros A, Cantor A, Scher J, Clemente J. Meta-Analysis of Trans-Disease Microbial Biomarkers of Protection and Pathogenesis in Autoimmune Conditions: Results from the AMP AIM Consortium [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/meta-analysis-of-trans-disease-microbial-biomarkers-of-protection-and-pathogenesis-in-autoimmune-conditions-results-from-the-amp-aim-consortium/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/meta-analysis-of-trans-disease-microbial-biomarkers-of-protection-and-pathogenesis-in-autoimmune-conditions-results-from-the-amp-aim-consortium/
