Session Information
Date: Wednesday, November 15, 2023
Title: Abstracts: Muscle Biology, Myositis & Myopathies – Basic & Clinical Science II
Session Type: Abstract Session
Session Time: 9:00AM-10:30AM
Background/Purpose: Sporadic inclusion body myositis (sIBM) is a subset of autoimmune inflammatory myopathies (AIM) that is often challenging to diagnose. The objective of this study was to use machine learning and a comprehensive profile of autoantibodies to: 1) examine sIBM endotypes, 2) differentiate sIBM from AIM. The primary goal was to identify an approach that would facilitate earlier and more precise diagnosis.
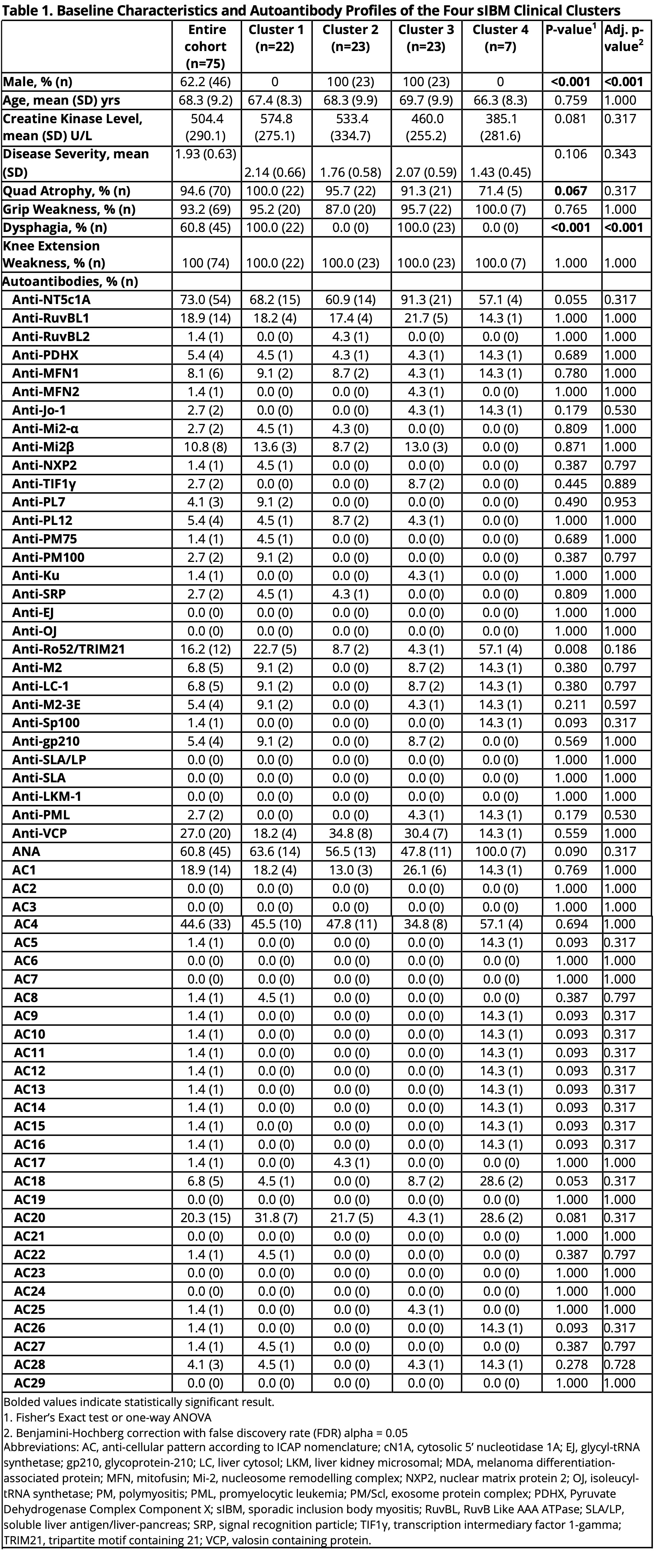
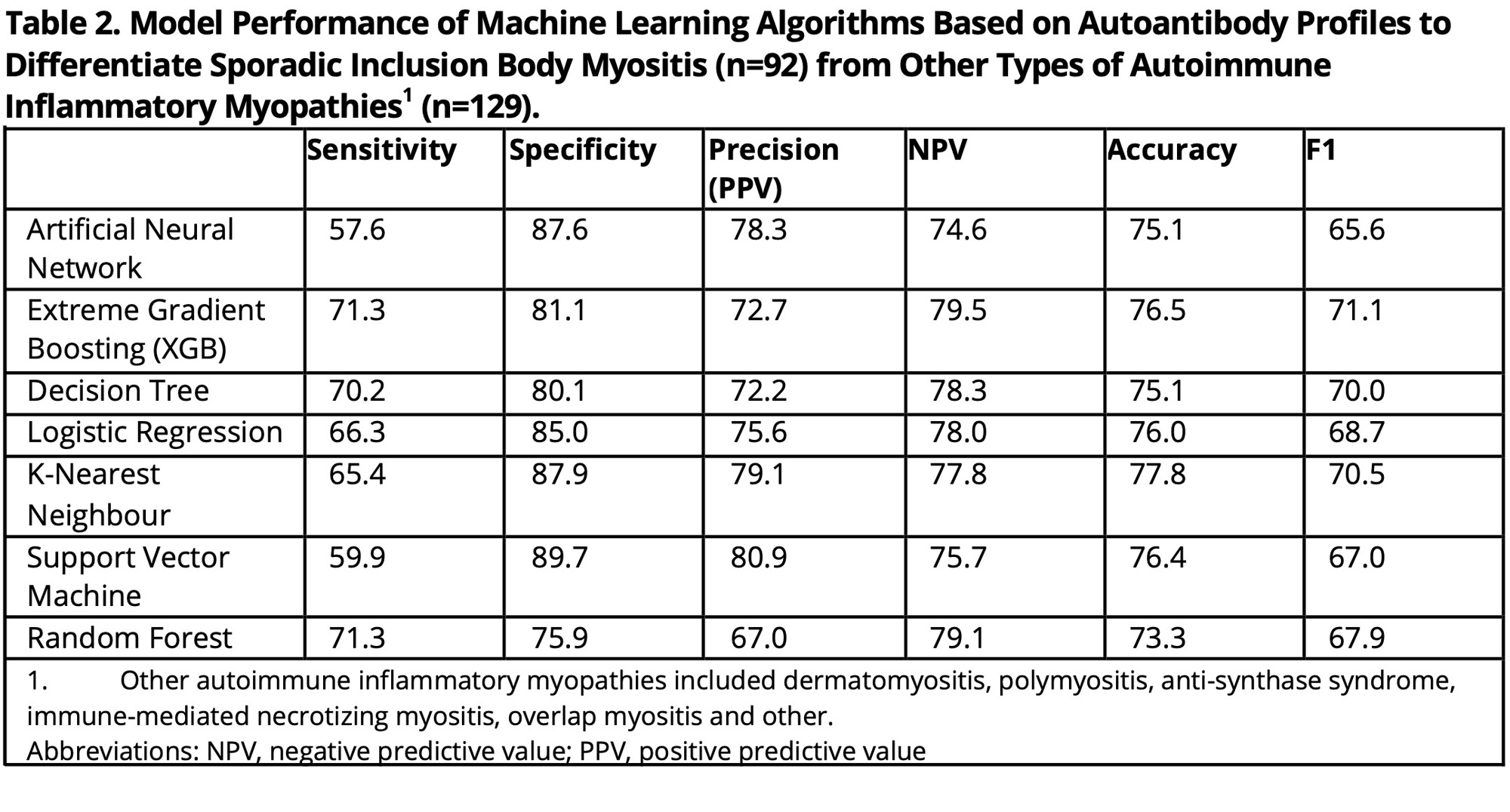
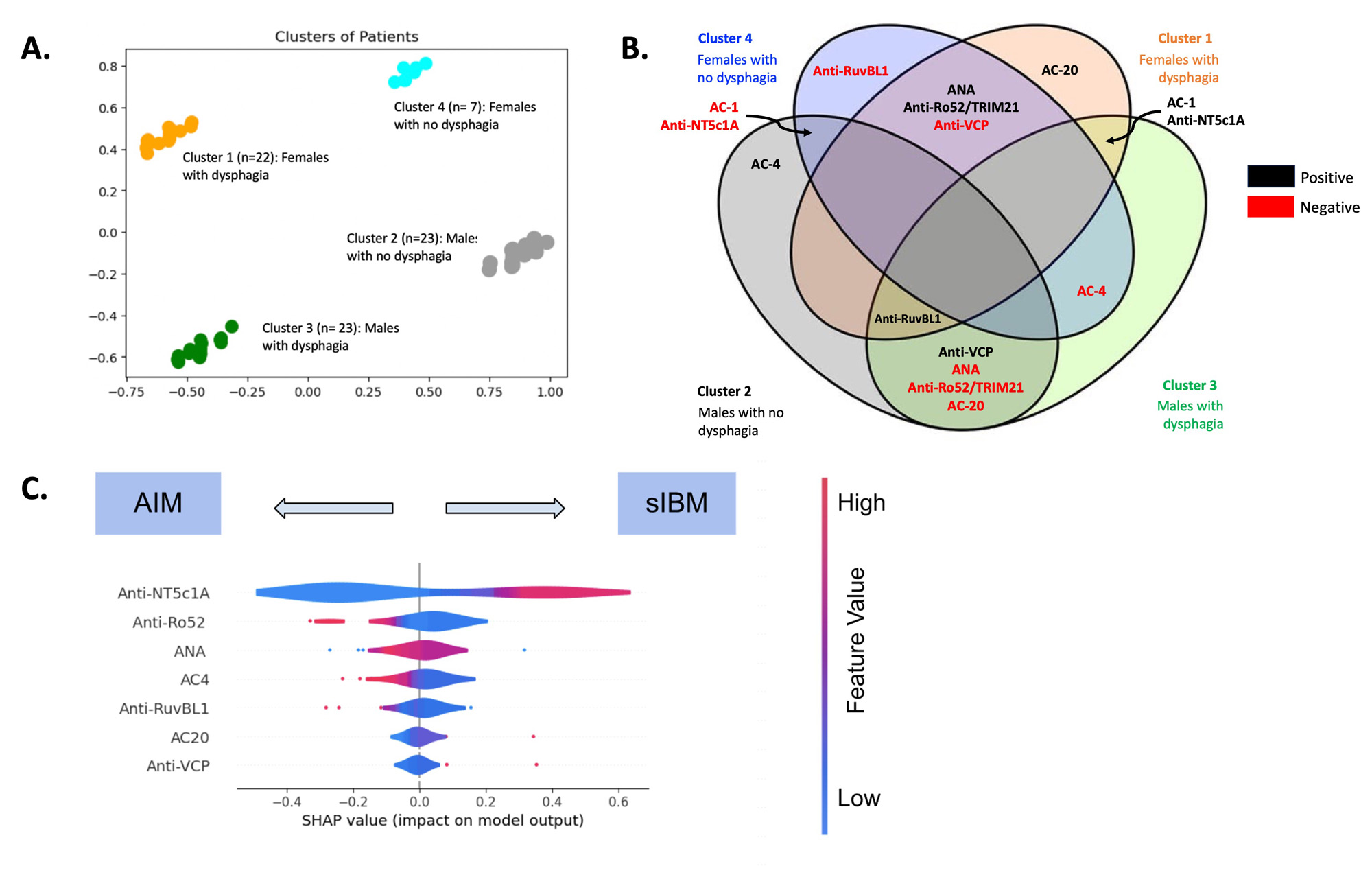
Methods: Clinical and demographic information were obtained on sIBM patients and AIM disease comparators. Baseline sera from these patients were tested for conventional and novel autoantibodies that included anti-NT5c1A/Mup44 and anti-mitofusin (MFN)-1 and -2) using an addressable laser bead immunoassay, a multiplexed autoimmune liver disease array, and antinuclear antibodies (ANA) on HEp-2 substrates by an indirect immunofluorescence assay (IFA). Agglomerative hierarchical clustering based on clinical features (creatine kinase level, dysphagia, knee extension weakness, quadriceps atrophy, grip strength, and disease severity (graded by expert)) was performed to identify clinical sub-phenotypes of sIBM. Seven classification algorithms were trained using K-fold cross validation to differentiate sIBM from AIM based on the autoantibodies and performance was assessed. SHapley Additive exPlanation (SHAP) plots identified the most important associated with each sIBM cluster and model used to different between sIBM vs. AIM.
Results: 75 sIBM patients were included in the cluster analysis (mean age 68.3 years (SD 9.2), 62.2% male) (Table 1). Four sIBM clinical clusters were identified: 1) females with dysphagia (n=22), 2) males with no dysphagia (n=23), 3) males with dysphagia (n=23), 4) females with no dysphagia (n=7) (Figure 1a). Based on the SHAP analysis, ANA and anti-Ro52/TRIM21 were more likely to be positive among female clusters (1 and 4), while anti-VCP was more likely to be positive among male clusters (2 and 3) (Figure 1b). Positive AC-1 (homogeneous pattern) and anti-NT5c1A were more likely to be found in clusters with dysphagia (1 and 3). Anti-RuvBL1 was less likely to be found among females with no dysphagia (cluster 4). Machine learning analysis based on autoantibody profile alone included an additional 17 sIBM patients (total 92) sIBM and 129 AIM disease controls. All the machine learning models used to differentiate sIBM vs. AIM, based on autoantibody profile alone, demonstrated high specificity (75.9%-89.7%) with fair sensitivity (57.6%-71.3%), accuracy (73.3%-77.8%) and F1 score (65.6%-71.1%) (Table 2). SHAP analysis revealed that positive anti-NT5c1A, AC-20 (cytoplasmic [speckled]) pattern, anti-VCP, but negative anti-Ro52/TRIM21, ANA, AC-4 (nuclear [fine speckled]) pattern, and anti-RuvBL1, favored a diagnosis of sIBM over AIM (Figure 1c).
Conclusion: In this comprehensive machine learning analysis of autoantibodies, ANA, anti-NT5c1A, anti-Ro52/TRIM21, anti-VCP, and anti-RuvBL1 were important biomarkers for both assessing sIBM clinical endotypes and differentiating sIBM from other AIMs. Future studies to study other novel biomarkers and validate our findings in larger cohorts are needed.
To cite this abstract in AMA style:
Wei J, Tarnopolsky M, Hudson M, Mitchell R, Buhler K, Dufour A, de Almeida L, Fortin P, Boilard E, Becker Y, Hatcher E, Zhang M, Fritzler M, Choi M. Machine Learning Identifies New Sporadic Inclusion Body Myositis Endotypes Associated with Unique Autoantibody Profiles [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/machine-learning-identifies-new-sporadic-inclusion-body-myositis-endotypes-associated-with-unique-autoantibody-profiles/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-identifies-new-sporadic-inclusion-body-myositis-endotypes-associated-with-unique-autoantibody-profiles/