Session Information
Date: Sunday, October 26, 2025
Title: Abstracts: Systemic Sclerosis & Related Disorders – Clinical I (0843–0848)
Session Type: Abstract Session
Session Time: 3:30PM-3:45PM
Background/Purpose: Systemic Sclerosis (SSc) is a clinically and molecularly heterogeneous autoimmune disease. We identified five intrinsic molecular subtypes in SSc by applying semi-supervised machine learning methods to multiple transcriptomic cohorts. A supervised single-sample classifier was developed for subtype predictions. Here, this predictive classifier is validated in multiple independent datasets and is used to comprehensively assess the association between molecular heterogeneity of SSc and key clinical features, extending our previous SSc subtyping research.
Methods: We trained a stacked‐ensemble model for intrinsic subtype prediction using GSVA enrichment scores in an integrated discovery cohort of 137 SSc and 37 Healthy participants (GSE9285, GSE32413, and GSE59787). Base learners were binary one-vs-all logistic regression models whose predicted probabilities were then combined by a Random Forest meta-learner. This model was applied to external DNA microarray and RNA-seq cohorts for assessment and validation. We tested associations between predicted subtypes and clinical features, such as FVC, DLCO, MRSS, ILD-risk, and autoantibody status. Discrete variables were compared with Odds Ratios (OR) test; continuous variables were tested using Wilcoxon rank-sum tests.
Results: We identified 5 intrinsic molecular subtypes of SSc through semi-supervised clustering and comparison to original publications. We identified an inflammatory-fibroproliferative group and an intermediate group between inflammatory and normal-like, which may represent a transitional subtype (Fig. 1A). Gene sets were identified that were most predictive for each subtype (Fig. 1B). We used the three broad subtype labels for classification due to small sample numbers. The best models showed AUROC of 0.94, 0.90, and 0.85 for inflammatory, normal-like, and fibroproliferative subtypes, respectively (Fig. 1C). The final, three-class model is able to predict SSc subtypes in independent DNA microarray and RNA-seq cohorts with strong concordance to the discovery set (Fig. 1D-E).Inflammatory patients were enriched for dcSSc (p < 0.001), had shorter disease duration (p < 0.001), and the highest MRSS (p < 0.001), exhibited increased ILD risk (p< 0.01) with reduced FVC/DLCO (p < 0.01), and were most likely to have RNA-polymerase III autoantibodies (p < 0.01)(Fig. 2). In contrast, Normal-like patients were enriched for lcSSc (OR ≈3.0, p< 0.001), late-stage disease (p < 0.01), longer disease duration (p < 0.001), lowest MRSS (p < 0.001), showed reduced ILD risk (p < 0.01) and preserved lung function (p < 0.01), and were most likely to carry ACA (p < 0.01)(Fig. 2). An intermediate MRSS and pulmonary impairment were observed in Fibroproliferative patients (p < 0.05). These subtype-clinical patterns were largely recapitulated in 4 additional, independent cohorts (Hinchcliff 8 plex, PRESS, GENISOS, ASSET) (Fig. 3).
Conclusion: Our second-generation classifier robustly predicts intrinsic SSc subtypes across studies and platforms. Each subtype shows consistent, biologically meaningful associations with disease phenotypes, severity, and autoantibody profiles.
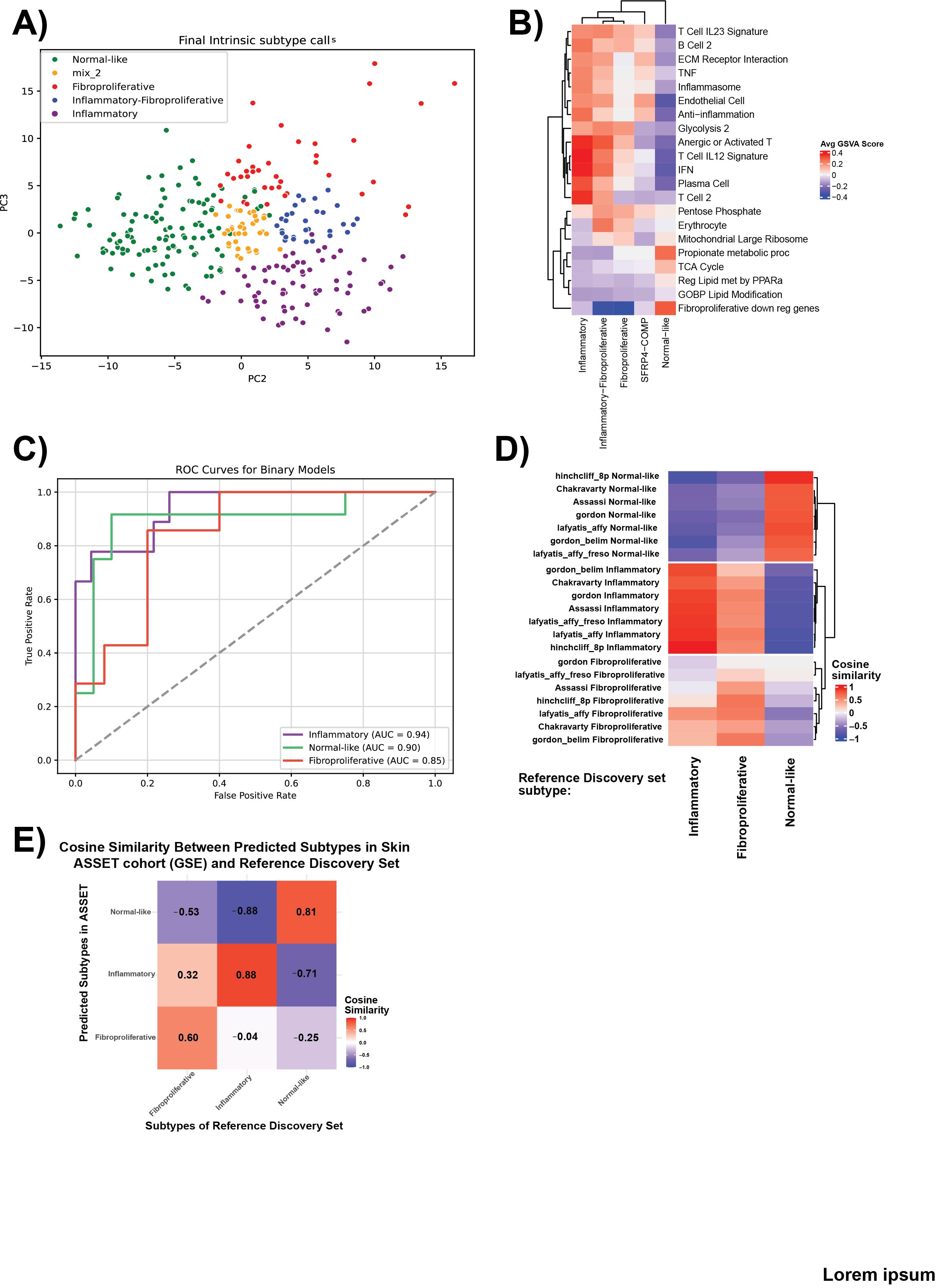 Figure 1. Identification and prediction of SSc intrinsic subtypes using semi-supervised and supervised machine learning approaches. (A) Semi-supervised constrained k-means clustering in the integrated discovery cohort (MPH, n = 174) defines five intrinsic subtypes and refines the “mixed” group into “inflammatory-fibroproliferative” and “intermediate” (between normal-like and inflammatory) subtypes. Principal component analysis (PCA) of all discovery samples colored by the final three broad subtype calls (inflammatory, normal-like, fibroproliferative) demonstrates clear separation.
Figure 1. Identification and prediction of SSc intrinsic subtypes using semi-supervised and supervised machine learning approaches. (A) Semi-supervised constrained k-means clustering in the integrated discovery cohort (MPH, n = 174) defines five intrinsic subtypes and refines the “mixed” group into “inflammatory-fibroproliferative” and “intermediate” (between normal-like and inflammatory) subtypes. Principal component analysis (PCA) of all discovery samples colored by the final three broad subtype calls (inflammatory, normal-like, fibroproliferative) demonstrates clear separation.
(B) Heatmap showing the up- and down-regulated pathways for the identified subtype. (C) Receiver operating characteristic (ROC) curves for one-vs-rest classifiers on a 20% hold-out set yield AUROC = 0.94 (inflammatory), 0.90 (normal-like), and 0.85 (fibroproliferative).
(D) Concordance of predicted subtypes in independent validation cohorts (DNA microarray) versus discovery labels, shown as confusion matrices and similarity metrics. (E) Concordance of predicted subtypes in ASSET RNA-seq cohort versus discovery reference samples.
.jpg) Figure 2. Clinical feature associations of intrinsic SSc molecular subtypes in MPH discovery set. (A) Disease duration (months) by subtype, shown as boxplots with Wilcoxon rank-sum p-values above each pairwise comparison.
Figure 2. Clinical feature associations of intrinsic SSc molecular subtypes in MPH discovery set. (A) Disease duration (months) by subtype, shown as boxplots with Wilcoxon rank-sum p-values above each pairwise comparison.
(B) Forest plot of odds ratios (95% CI) for diffuse cutaneous versus limited cutaneous SSc (dcSSc vs. lcSSc) for each subtype, relative to Normal-like (dashed vertical line at OR=1).
(C) Modified Rodnan skin score (MRSS) by subtype, with p-values from Wilcoxon tests.
(D–G) Forest plots of odds ratios (95% CI) for serologic and pulmonary categorical features: (D) RNA-polymerase III autoantibody, (E) anti-centromere autoantibody, (F) ANA positivity, and (G) interstitial lung disease (ILD) presence.
(H–J) Pulmonary function measures (% predicted) by subtype—(H) FEV₁, (I) FVC, and (J) total lung capacity (TLC)—shown as boxplots with Wilcoxon test p-values.
Subtype colors are fibroproliferative (red), inflammatory (purple), normal-like (green), and intermediate (yellow).
.jpg) Figure 3. Validation of subtype–clinical associations in independent cohorts.
Figure 3. Validation of subtype–clinical associations in independent cohorts.
(A–B) Disease duration (months) by molecular subtype in the Hinchliff microarray (A) and PRESS RNA-seq cohort (B), shown as boxplots with Wilcoxon rank-sum p-values.
(C–D) Forest plots of odds ratios (95% CI) for diffuse versus limited cutaneous SSc (dcSSc vs lcSSc) by subtype in Hinchliff (C) and GENISOS (D), relative to Normal-like (dashed line at OR = 1). (E–H) MRSS by subtype in Hinchliff (E), GENISOS (F), PRESS (G), and ASSET baseline (H), with p-values from Wilcoxon tests. (I–K) Odds ratios (95% CI) for RNA-polymerase III autoantibody positivity by subtype in Hinchliff (I), PRESS (J), and ASSET (K).
(L) Odds ratio (95% CI) for RNA-polymerase I autoantibody positivity by subtype in ASSET.
Subtype color key: fibroproliferative (red), inflammatory (purple), normal-like (green), intermediate/other (yellow).
To cite this abstract in AMA style:
Gong Z, Parvizi R, Jarnagin H, Chen H, Morrisson M, Wood T, Hinchcliff M, Khanna D, Whitfield M. Machine Learning–Based Skin Transcriptome Classifier (v2.0) Links SSc Molecular Subtypes to Disease Severity and Progression [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/machine-learning-based-skin-transcriptome-classifier-v2-0-links-ssc-molecular-subtypes-to-disease-severity-and-progression/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-based-skin-transcriptome-classifier-v2-0-links-ssc-molecular-subtypes-to-disease-severity-and-progression/
