Session Information
Date: Monday, November 8, 2021
Session Type: Poster Session C
Session Time: 8:30AM-10:30AM
Background/Purpose: Few studies on rheumatoid arthritis (RA) have generated machine learning models to predict biologic disease-modifying antirheumatic drugs (bDMARDs) responses; however, these studies included insufficient analysis on important features. Moreover, machine learning is yet to be used to predict bDMARD responses in ankylosing spondylitis (AS). Thus, in this study, machine learning was used to predict such responses in RA and AS patients.
Methods: Data were retrieved from the Korean College of Rheumatology Biologics therapy (KOBIO) registry. The number of RA and AS patients in the training dataset were 625 and 611, respectively. We prepared independent test datasets that did not participate in any process of generating machine learning models. Baseline clinical characteristics were used as input features. Responders were defined as those who met the ACR 20% improvement response criteria (ACR20) and ASAS 20% improvement response criteria (ASAS20) in RA and AS, respectively, at the first follow-up. Multiple machine learning methods, including random forest (RF), were used to generate models to predict bDMARD responses, and we compared them with the logistic regression model.
Results: The RF model had superior prediction performance to logistic regression model (accuracy: 0.726 [95% confidence interval (CI): 0.725–0.730] vs. 0.689 [0.606–0.717], area under curve (AUC) of the receiver operating characteristic curve (ROC) 0.638 [0.576–0.658] vs. 0.565 [0.493–0.605], F1 score 0.841 [0.837–0.843] vs. 0.803 [0.732–0.828], AUC of the precision-recall curve 0.808 [0.763–0.829] vs. 0.754 [0.714–0.789]) with independent test datasets in patients with RA (Figure 1). However, machine learning and logistic regression exhibited similar prediction performance in AS patients. Furthermore, the patient self-reporting scales, which are patient global assessment of disease activity (PtGA) in RA and Bath Ankylosing Spondylitis Functional Index (BASFI) in AS, were revealed as the most important features in both diseases (Figure 2).
Conclusion: RF exhibited superior prediction performance for responses of bDMARDs to a conventional statistical method, i.e., logistic regression, in RA patients. In contrast, despite the comparable size of the dataset, machine learning did not outperform in AS patients. The most important features of both diseases, according to feature importance analysis were patient self-reporting scales.
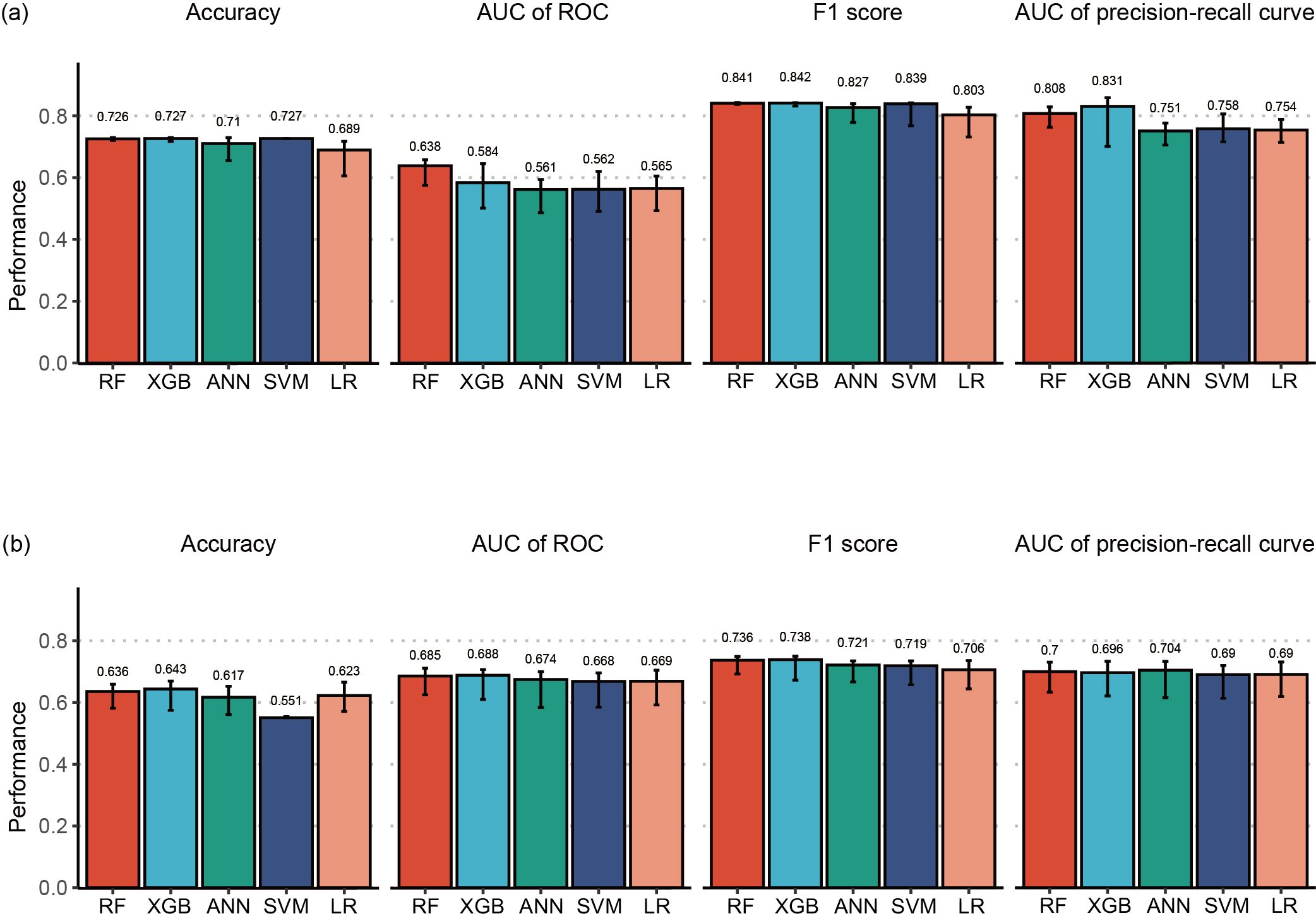 Figure 1. Performances of models trained using various methods (RF, XGBoost, ANN, SVM, and logistic regression) with independent test dataset, in terms of accuracy, AUC of the ROC curve, F1 score, and AUC of the precision-recall curve. (a) RA patients. (b) AS patients. RF, random forest; XGBoost, extreme gradient boosting; ANN, artificial neural network; SVM, support vector machine; LR, logistic regression.
Figure 1. Performances of models trained using various methods (RF, XGBoost, ANN, SVM, and logistic regression) with independent test dataset, in terms of accuracy, AUC of the ROC curve, F1 score, and AUC of the precision-recall curve. (a) RA patients. (b) AS patients. RF, random forest; XGBoost, extreme gradient boosting; ANN, artificial neural network; SVM, support vector machine; LR, logistic regression.
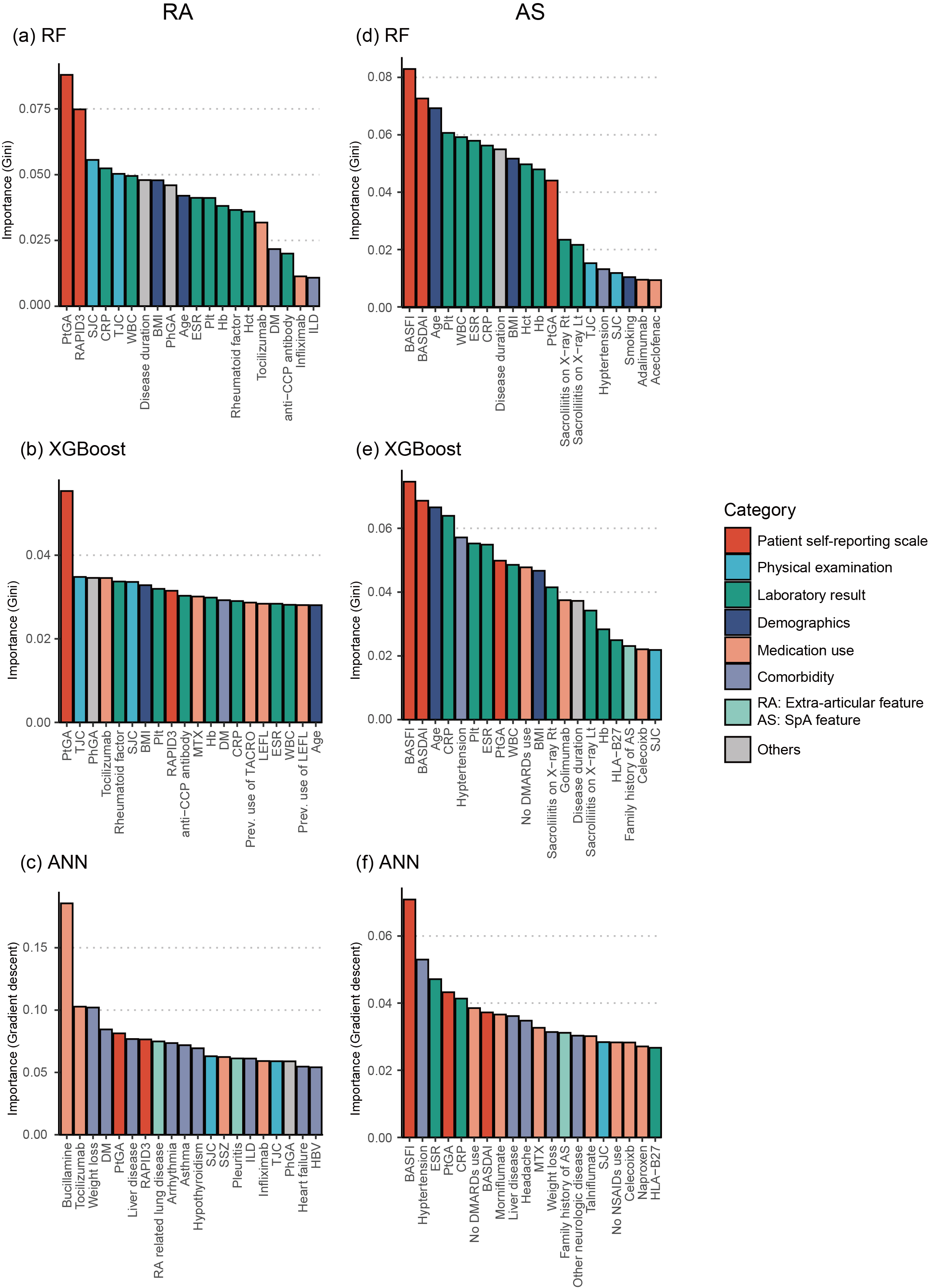 Figure 2. Result of feature importance analysis from the best performing models of each machine learning method. The X-axis represents the input clinical features. The Y-axis represents the feature importance score calculated using the Gini importance or risk backpropagation methods in RF/XGBoost and ANN, respectively. The color of columns represents the categories in which the feature was included. Top 20 important features are shown in figures. Feature importance of (a) RF model, (b) XGBoost model, and (c) ANN model in patients with RA. Feature importance of (d) RF model, (e) XGBoost model, and (f) ANN model in patients with AS. WBC, white cell count; BMI, body mass index; Plt, platelet; Hb, hemoglobin; Hct, hematocrit; DM, diabetes mellitus; anti-CCP, anti-cyclic citrullinated protein; ILD, interstitial lung disease; MTX, methotrexate; TACRO, tacrolimus; LEFL, leflunomide; SSZ, sulfasalazine.
Figure 2. Result of feature importance analysis from the best performing models of each machine learning method. The X-axis represents the input clinical features. The Y-axis represents the feature importance score calculated using the Gini importance or risk backpropagation methods in RF/XGBoost and ANN, respectively. The color of columns represents the categories in which the feature was included. Top 20 important features are shown in figures. Feature importance of (a) RF model, (b) XGBoost model, and (c) ANN model in patients with RA. Feature importance of (d) RF model, (e) XGBoost model, and (f) ANN model in patients with AS. WBC, white cell count; BMI, body mass index; Plt, platelet; Hb, hemoglobin; Hct, hematocrit; DM, diabetes mellitus; anti-CCP, anti-cyclic citrullinated protein; ILD, interstitial lung disease; MTX, methotrexate; TACRO, tacrolimus; LEFL, leflunomide; SSZ, sulfasalazine.
To cite this abstract in AMA style:
Lee S, Kang S, Eun Y, Kim H, Lee J, Koh E, Cha H. Machine Learning Based Prediction Model for Responses of bDMARDs in Patients with Rheumatoid Arthritis and Ankylosing Spondylitis [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/machine-learning-based-prediction-model-for-responses-of-bdmards-in-patients-with-rheumatoid-arthritis-and-ankylosing-spondylitis/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-based-prediction-model-for-responses-of-bdmards-in-patients-with-rheumatoid-arthritis-and-ankylosing-spondylitis/
