Session Information
Date: Monday, October 27, 2025
Title: (0955–0977) Systemic Sclerosis & Related Disorders – Basic Science Poster I
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Clinically useful biomarkers for systemic sclerosis (SSc) are needed. While obtaining samples from affected organs like the skin and lungs is invasive and cannot be done routinely, serum is collected for routine clinical care. Prior studies showed serum protein profiles align more with skin molecular changes than with peripheral blood cells (PBCs), though they analyzed a limited number of proteins. The current study characterizes longitudinal large-scale serum proteomic profile alongside global gene expression in PBCs and skin biopsies from SSc patients and healthy controls.
Methods: Three-level samples were collected from 109 SSc patients (disease duration < 6 year) and 42 matched controls. 98 SSc patients had a second sample and 47 had a third sample. 5416 serum proteins were measured using Olink Explore HT panel in serum samples. In addition, bulk global gene expression profiling was performed on concurrently collected skin and PBC RNA samples. An FDR adjusted p-value of 0.05 was considered statistically significant.
Results: The mean disease duration was 2.71 years and 61% of patients had diffuse SSc. In comparison of baseline SSc samples to controls, 697 proteins were significantly differentially expressed. Moreover, 249 proteins significantly correlated with modified Rodnan Skin Score (mRSS) (Figure 1A), of which 184 proteins were also differentially expressed in concordant direction in SSc vs control comparison (Figure 1B), indicating a significant correlation between direction of differential expression of serum proteins in SSc vs. control comparison and their association with mRSS (Figure 1C – permutation p-value < 0.0001 – Figure 1D). The differential expression of serum proteins in SSc vs. control comparison correlated more strongly with the differential expression of corresponding transcripts in skin than in PBCs (ρ=0.17 vs ρ=0.081, Figure 2). Similar to previously reported observations in skin, a comparison of the last to the initial serum samples in patients with early diffuse disease (n=36 – disease duration < 3 years) showed that the follow up serum samples clustered nearer to healthy controls than did initial serum samples (see the PCA analysis shown in Figure 3). Similar results were found when the skin transcriptomic and serum proteomic data were integrated using Similarity Network Fusion (SNF) analysis.Among differentially expressed serum proteins, 141 baseline protein levels (20.1%) showed predictive significance for the rate of longitudinal change in mRSS after adjustment for disease duration and gender among patients with diffuse systemic sclerosis (Figure 3C), with 23 proteins being associated with faster increases in mRSS while 118 proteins were associated with slower increase or even a decrease in mRSS over time.
Conclusion: SSc proteomic signature reflects more closely the molecular dysregulations of skin than PBC and shows a trend towards normalization, paralleling the findings at the skin transcriptomic level. Moreover, serum proteins are associated with rate of change in mRSS over time and might aid in identifying patients with rapidly progressing skin fibrosis.
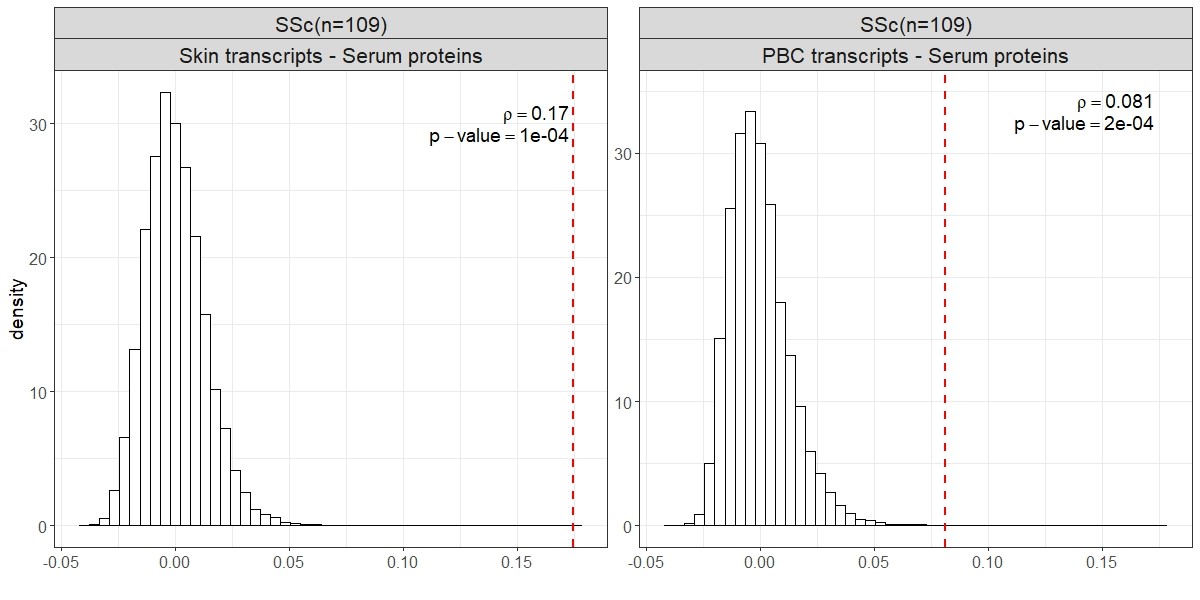 Figure 1. A) Volcano plot of differences between SSc-Controls; B) Volcano plot of the correlation between proteins and mRSS in SSc; C) Scatter plot of associations with mRSS and SSc-Controls differences; D) Permutation-based significance of correlation between differential serum protein expression (SSc vs. control) and its association with mRSS. The vertical red dashed line indicates the observed Spearman correlation coefficient based on the original sample annotation, while the histogram represents the null distribution of correlation values obtained from randomly permuted sample labels.
Figure 1. A) Volcano plot of differences between SSc-Controls; B) Volcano plot of the correlation between proteins and mRSS in SSc; C) Scatter plot of associations with mRSS and SSc-Controls differences; D) Permutation-based significance of correlation between differential serum protein expression (SSc vs. control) and its association with mRSS. The vertical red dashed line indicates the observed Spearman correlation coefficient based on the original sample annotation, while the histogram represents the null distribution of correlation values obtained from randomly permuted sample labels.
.jpg) Figure 2. Concordance of between-sample similarities, measured by Spearman correlations, for pairwise comparisons of protein expression between skin and PBC. The Mantel test’s observed concordance (vertical red dashed lines) was evaluated against null distributions created by random permutation of samples within each dataset.
Figure 2. Concordance of between-sample similarities, measured by Spearman correlations, for pairwise comparisons of protein expression between skin and PBC. The Mantel test’s observed concordance (vertical red dashed lines) was evaluated against null distributions created by random permutation of samples within each dataset.
.jpg) Figure 3. A) Principal component analysis (PCA) with differentially expressed serum proteins in healthy controls, first and last biopsies from early diffuse patients. B) Similarity Network Fusion (SNF) analysis of longitudinal serum protein and skin gene expression data from healthy controls and from the initial and final biopsies of early diffuse SSc patients. C) Volcano plot of serum proteins predictive of longitudinal mRSS change.
Figure 3. A) Principal component analysis (PCA) with differentially expressed serum proteins in healthy controls, first and last biopsies from early diffuse patients. B) Similarity Network Fusion (SNF) analysis of longitudinal serum protein and skin gene expression data from healthy controls and from the initial and final biopsies of early diffuse SSc patients. C) Volcano plot of serum proteins predictive of longitudinal mRSS change.
To cite this abstract in AMA style:
Choi B, Pedroza C, Skaug B, Mayes M, Assassi S. Longitudinal Multiomic Study of Skin, Peripheral Blood, and Serum: Serum Proteome Reflects the Disease Process at the End-organ Level and Predicts the Course of Modified Rodnan Skin Score [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/longitudinal-multiomic-study-of-skin-peripheral-blood-and-serum-serum-proteome-reflects-the-disease-process-at-the-end-organ-level-and-predicts-the-course-of-modified-rodnan-skin-score/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/longitudinal-multiomic-study-of-skin-peripheral-blood-and-serum-serum-proteome-reflects-the-disease-process-at-the-end-organ-level-and-predicts-the-course-of-modified-rodnan-skin-score/
