Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: A loss of systemic self-tolerance to anti-nuclear autoantibodies (ANAs) is one of the main hallmarks of SLE. However, most healthy females with ANAs will never develop clinical illness. Incomplete lupus (ILE) patients exhibit some clinical symptoms, while most never progress to SLE. It is unknown what triggers ANA+ healthy individuals to progress to clinical illness. Alterations in B and T cell receptor repertoires (BCR/TCR) of patients with different autoimmune diseases have been found to affect immune tolerance. This study investigates changes in immune receptors repertoires in preclinical SLE.
Methods: We selected 64 individuals of African and European American ancestry (ANA-, ANA+ healthy controls, ILE, SLE). Single cell transcriptomics, VDJ-clone and surface protein profiles for PBMCs were developed using Single Cell Immune Profiling scRNA-Seq platform (10x Genomics) along with TotalSeq Human Universal cocktail (Biolegend) to assess B/T cell clonal expansion, identify VDJ gene usage during preclinical autoimmunity development and find transcriptionally similar clonotypes. BCR/TCR data were analyzed with Python packages Scirpy and Dandelion.
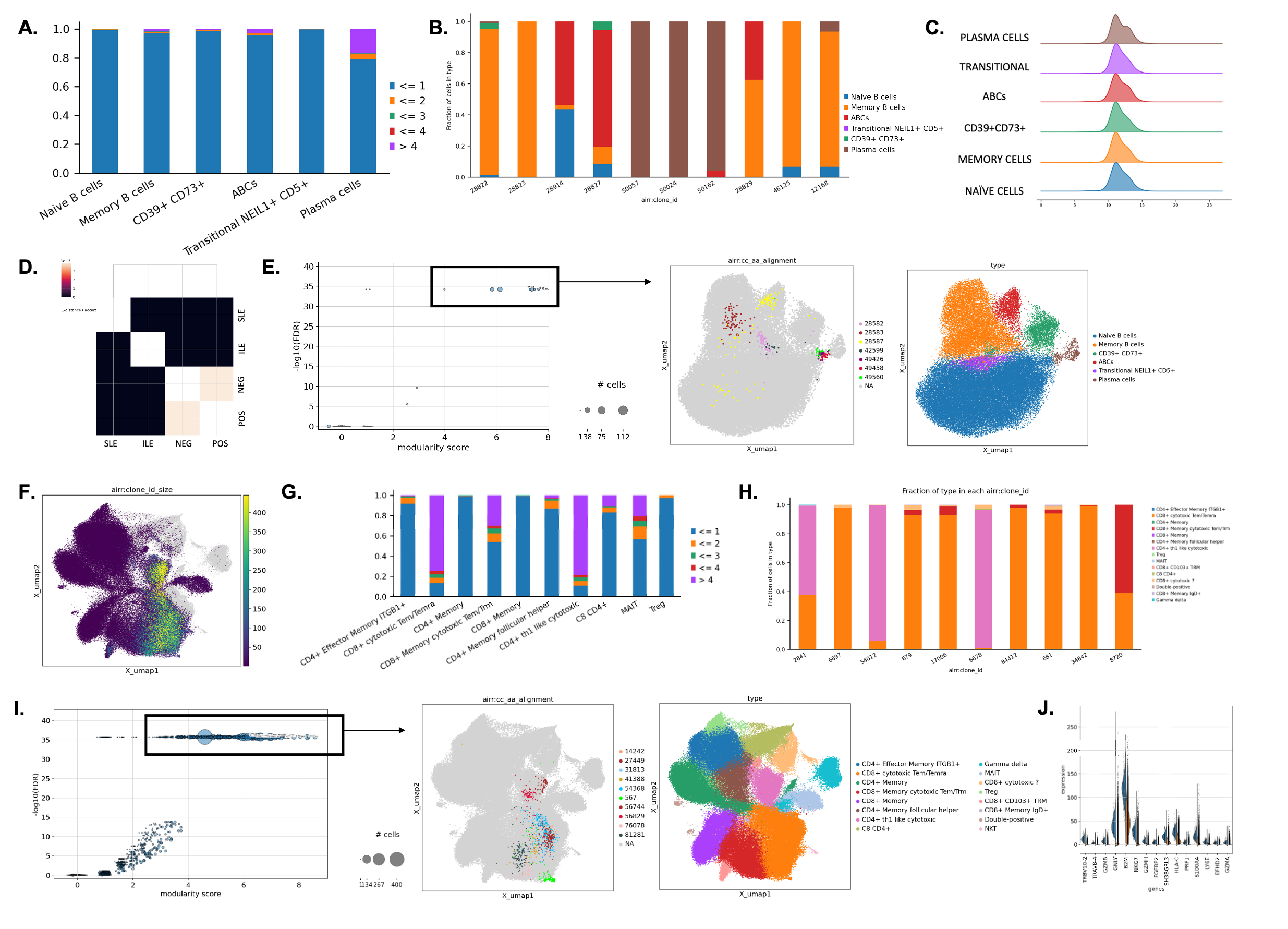
Results: We found higher number of expanded clonotypes in ILE and SLE individuals, particularly of Memory B cells, ABCs and Plasma cells (Fig.1A). It appears clonal expansion is present mostly in individuals of European American ancestry. The largest clonotypes are present across those three populations (Fig.1B). Additionally, we observed larger CDR3 lengths in ABCs and Plasma cells (Fig.1C). B cell receptor repertoire appear to have an overlap in ANA- and ANA+ healthy individuals; however not between disease groups (Fig.1D). Clonotypes with highest modularity score; indicating transcriptional similarity, are restricted to Memory B cells, ABCs and Plasma cell populations (Fig.1E). Differential expression analysis of B cells in the two top scoring clonotypes indicates involvement of the following genes: IGKV1-5, IGLV7-46, IGHV1-69, IGHG1, IGHV1-69D, as well as S100A8, IFI30. One of the most abundant V genes (IGKV2D) is mostly related to ANA+ individuals, while IGLV4-3 and IGKV2D-30 across SLE. We found cytotoxic T cells have largest fraction of expanded clonotypes with further increase in ANA+ individuals (Fig.1F-H). However, we haven’t observed differences in expansion by ancestry. Clonotypes with a high modularity score, consisting of cells with similar molecular phenotype, are located mostly within CD8+ cytotoxic T cells and CD4+ Th1-like populations and are associated with higher expression of TXNIP, TMSB4X, HLA, GZMB (Fig.1I-J).
Conclusion: In conclusion, we found specific clonal characteristics affecting preclinical autoimmunity development that suggest involvement of lymphocyte population expansion.
To cite this abstract in AMA style:
Bylinska A, Smith M, Lu R, Jones B, Guthridge C, Marlin M, Wright C, Macwana S, DeJager W, Beel M, Lessard C, Arriens C, Merrill J, James J, Guthridge J. Investigating Adaptive Immune Receptor Repertoires by Deep Immune Cell Phenotyping in Preclinical Autoimmunity Development [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/investigating-adaptive-immune-receptor-repertoires-by-deep-immune-cell-phenotyping-in-preclinical-autoimmunity-development/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/investigating-adaptive-immune-receptor-repertoires-by-deep-immune-cell-phenotyping-in-preclinical-autoimmunity-development/