Session Information
Date: Friday, November 6, 2020
Session Type: Abstract Session
Session Time: 5:00PM-5:50PM
Background/Purpose: Psoriatic Arthritis (PsA) is a progressive inflammatory arthritis that occurs in about 24% of psoriasis patients. Early diagnosis of PsA is associated with better outcomes. Since almost 90% of patients develop PsA either simultaneously or after the onset of cutaneous psoriasis, careful evaluation of psoriasis patients may increase early identification of PsA. Although several candidate protein markers have been demonstrated to be associated with PsA, individually the power to discriminate between PsA and psoriasis without PsA (PsC) is poor. Here, we have developed a computational approach to identify 9 alternative signatures by combining clinical and protein markers to improve discrimination between PsA and PsC.
Methods: Serum samples were obtained from 192 PsA and 191 PsC patients (Table 1). Sixteen protein markers identified in previous studies were assayed in serum samples using ELISA and combined with four clinical features associated with PsA and easily identified by non-rheumatologists (Table 1). To classify patients, we used Support Vector Machines (SVM). To identify the best performing signatures (markers and classifier parameters) we used a three-layer approach. In the first two layers, for training and testing classifiers, we used data only from patients with polyarticular PsA (tender or swollen joint count > 4), while in the third layer, we used all PsA samples. Evaluation of signatures was done using 10-fold cross-validation. First, using a greedy algorithm we identified 41 signatures with accuracy above 75%. Next, we identified 9 signatures using four different measures based on frequency of features and AUCs of these 41 signatures. In the third layer, we used three sample sets for training and testing 2700 classifiers: all samples, only oligoarticular (tender or swollen joint count ≤ 4) PsA, and only polyarticular PsA. All 9 classifiers were trained and tested on similar training and testing sets. Finally, we prioritized the 9 signatures based on their AUC ranks (Table 2).
Results: Demographics and clinical features of patients are provided in Table 1. We propose 9 signatures as alternative signatures to use for distinguishing PsA from PsC, depending on available data, all of which share four features: nail psoriasis, LEP, CRP, and SOST (Table 2). Importantly, the lowest ranked signatures were those without SPP1, suggesting its importance in classifying patients. Our top feature set (Table 2) obtained AUC of up-to 90% on polyarticular samples (median 75%). Performance slightly dropped when we considered all samples, or only oligoarticular samples (Figure 1). Median AUC of 68% (max AUC = 86%) for distinguishing oligoarticular PsA cases from PsC is significant considering high similarity between patients of these two groups.
Conclusion: Integration of clinical and biomarker data through machine learning improves discrimination of PsA from PsC patients. Nail psoriasis and CRP followed by SPP1 are the most important features influencing performance of classifiers. Nail psoriasis, CRP, DEFA1, LEP, SOST, SPP1, TFCP2_CPII, TNFRSF11B provided best discrimination between PsA and PsC and needs further validation.
 Table 1. Clinical and demographic information about samples
Table 1. Clinical and demographic information about samples
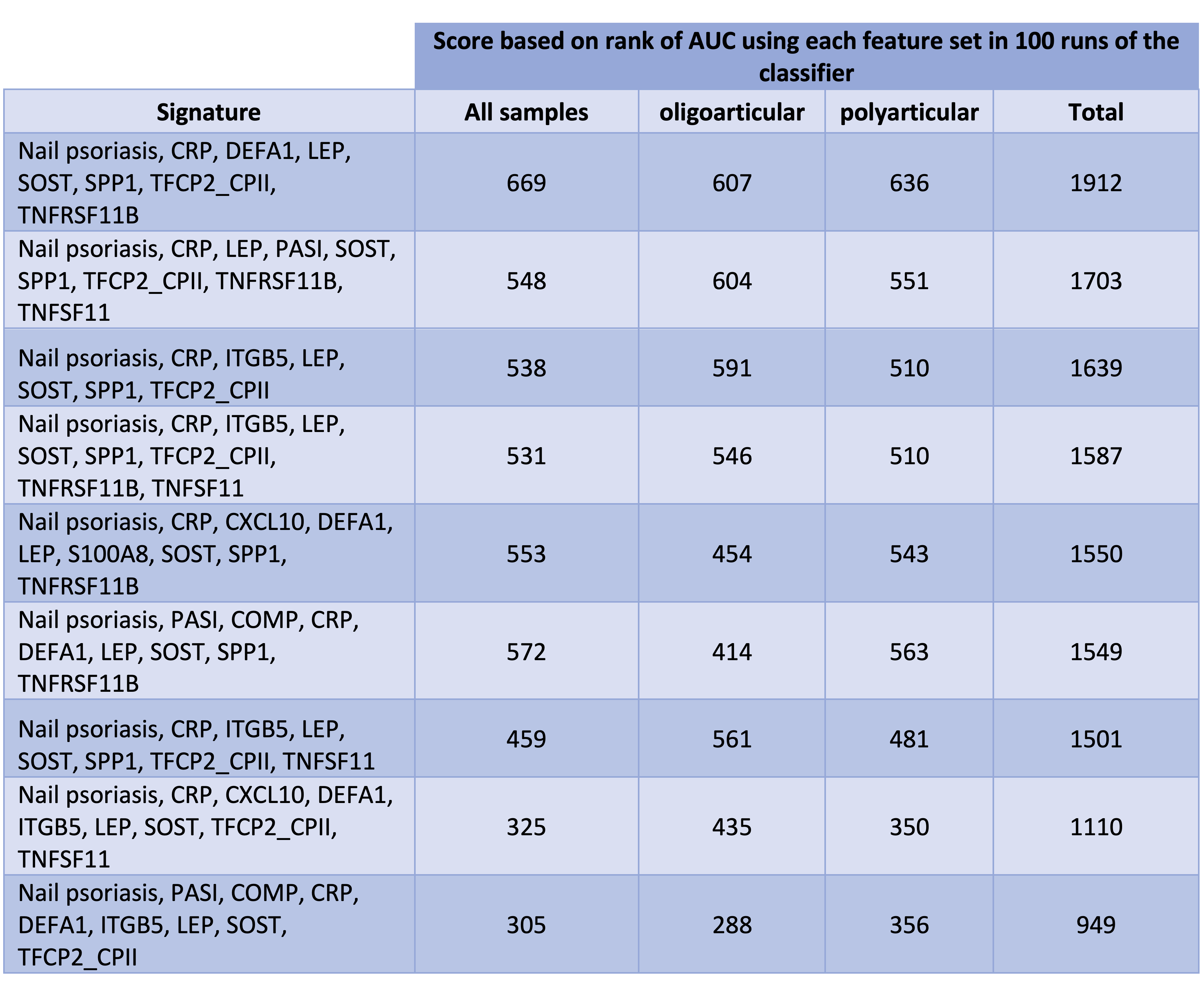 Table 2. 9 identified feature sets sorted based on their score (the higher the score, the better) We used same combinations of samples to train and test all 9 classifiers. To obtain the scores, for each training and testing set, we ranked all of the 9 classifiers from 1 to 9 (1 for the lowest and 9 for the highest AUC). Next, for each signature, we multiplied its frequency at each rank by the rank. The scores in this table are the sum up of these 9 values for each of the three categories of samples.
Table 2. 9 identified feature sets sorted based on their score (the higher the score, the better) We used same combinations of samples to train and test all 9 classifiers. To obtain the scores, for each training and testing set, we ranked all of the 9 classifiers from 1 to 9 (1 for the lowest and 9 for the highest AUC). Next, for each signature, we multiplied its frequency at each rank by the rank. The scores in this table are the sum up of these 9 values for each of the three categories of samples.
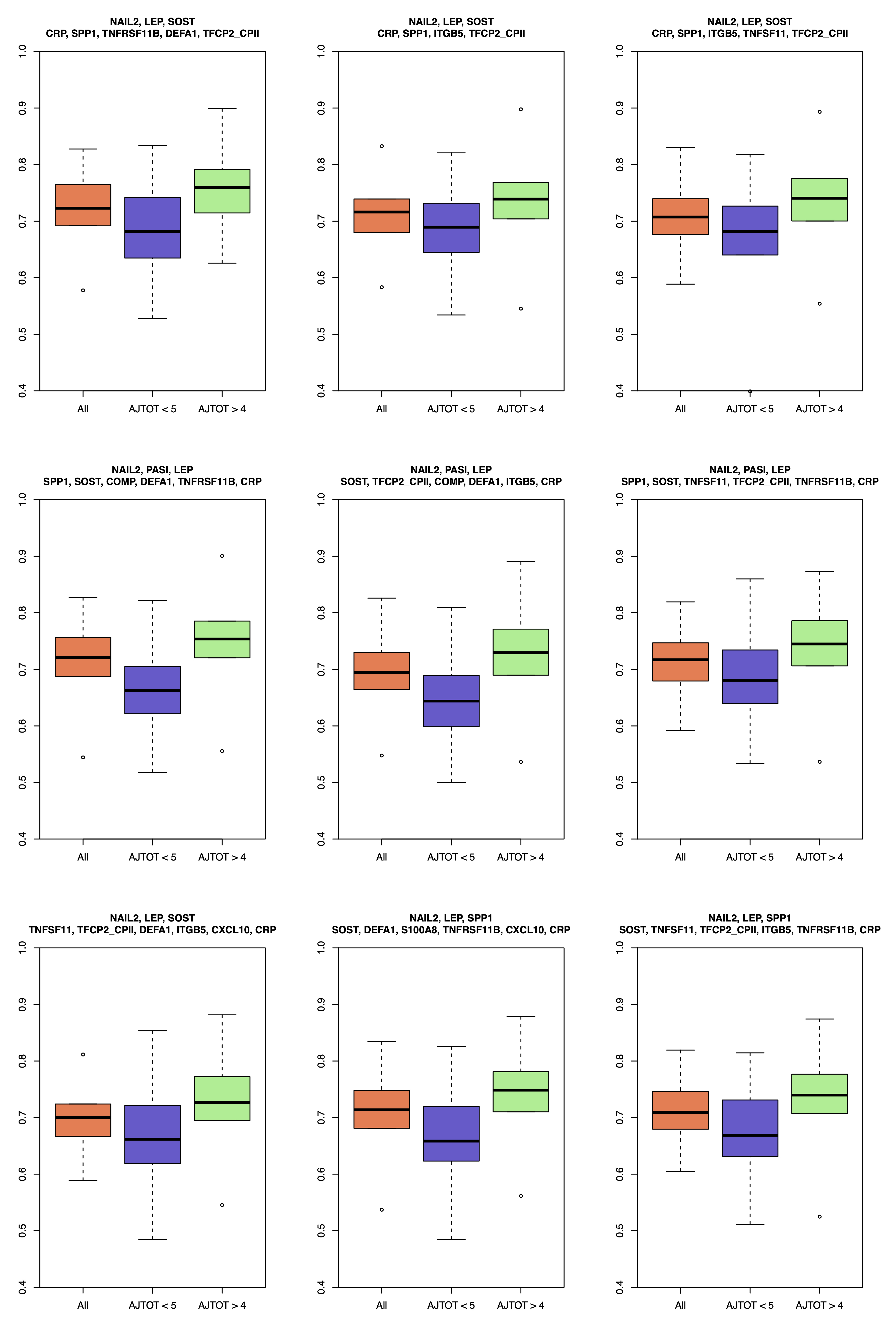 Figure 1. Distribution of AUC of SVMs (y-axis) using each of the 9 identified signatures (listed above each panel) as their feature sets. Each classifier was trained and tested 100 times using three different samples sets: all PsC samples and 1) only polyarticular samples; 2) only oligoarticular samples; 3) all samples. Three boxplots in each panel shows AUC distribution relevant to these three sample sets. For all 9 classifiers, similar training and testing sets were used, making their performance comparable. While all 9 signatures are among high performing signatures, the top left panel was the highest rank in our list.
Figure 1. Distribution of AUC of SVMs (y-axis) using each of the 9 identified signatures (listed above each panel) as their feature sets. Each classifier was trained and tested 100 times using three different samples sets: all PsC samples and 1) only polyarticular samples; 2) only oligoarticular samples; 3) all samples. Three boxplots in each panel shows AUC distribution relevant to these three sample sets. For all 9 classifiers, similar training and testing sets were used, making their performance comparable. While all 9 signatures are among high performing signatures, the top left panel was the highest rank in our list.
To cite this abstract in AMA style:
Rahmati S, Abji F, Rahman P, Chandran V. Integration of Clinical and Protein Markers Through Machine Learning to Distinguish Patients with Psoriasis Arthritis from Those with Psoriasis Without Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/integration-of-clinical-and-protein-markers-through-machine-learning-to-distinguish-patients-with-psoriasis-arthritis-from-those-with-psoriasis-without-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/integration-of-clinical-and-protein-markers-through-machine-learning-to-distinguish-patients-with-psoriasis-arthritis-from-those-with-psoriasis-without-psoriatic-arthritis/
