Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Multiple factors can predispose individuals to development of SLE, including the presence of African American ancestry, lupus-associated autoantibodies (ANAs), or some clinical manifestations of SLE, termed incomplete lupus (ILE). However, most ILE patients never progress to SLE and few ANA+ individuals develop autoimmune disease. We sought to identify protein signatures and cell populations involved in driving transition to full immune cell dysregulation.
Methods: Serum from 64 subjects with African or European ancestry, divided evenly among healthy individuals (ANA-), healthy with ANAs (ANA+), ILE, and SLE were examined using the Explore HT proximity extension assay (Olink) to measure the levels of >5400 proteins. Single cell transcriptomic, VDJ-clone, and surface protein profiles for PBMCs were developed using the Single Cell Immune Profiling scRNA-seq platform (10x Genomics) with the TotalSeq Human Universal Cocktail (Biolegend) to identify disease associated cell populations, differential gene signatures and dysregulated pathways.
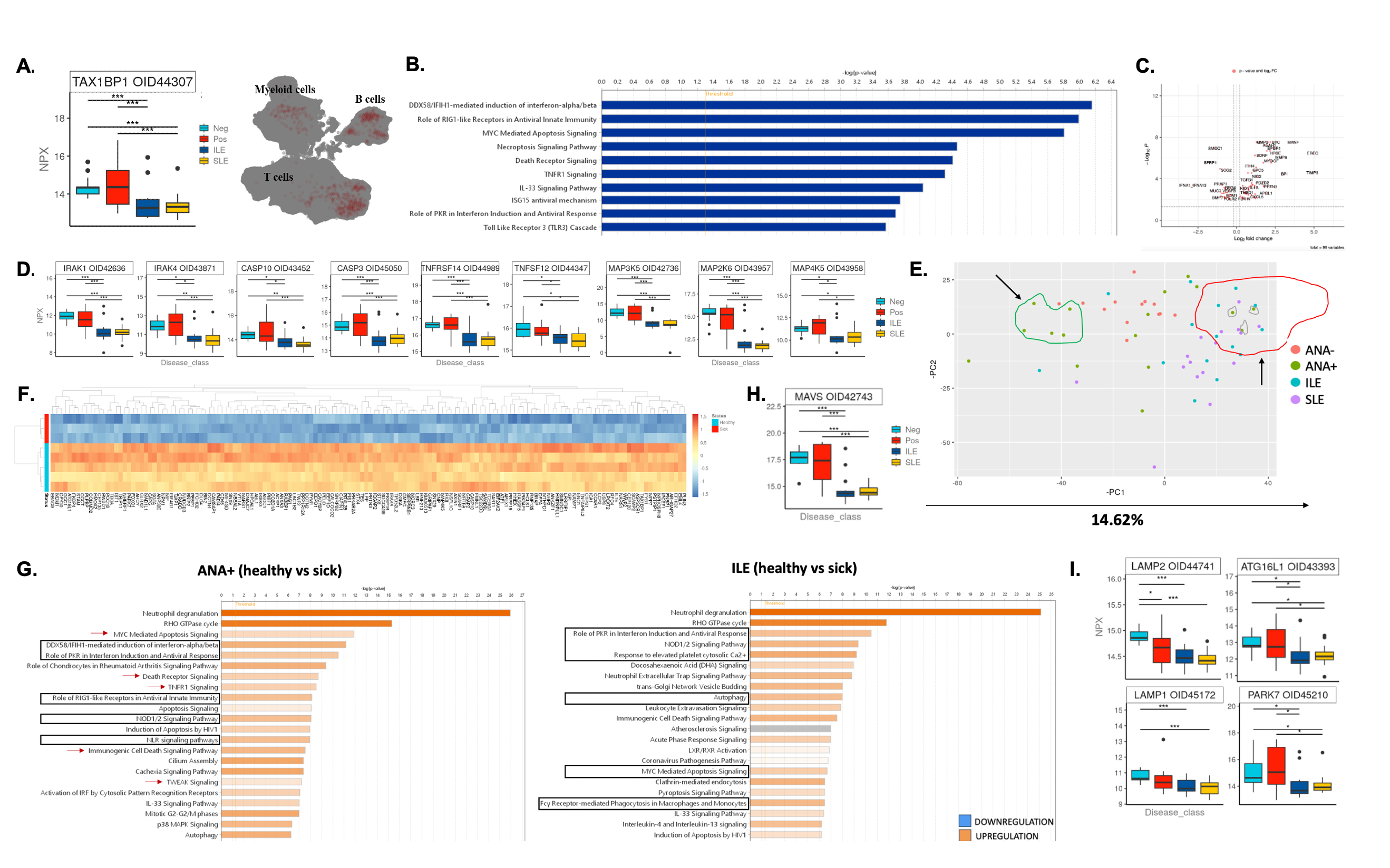
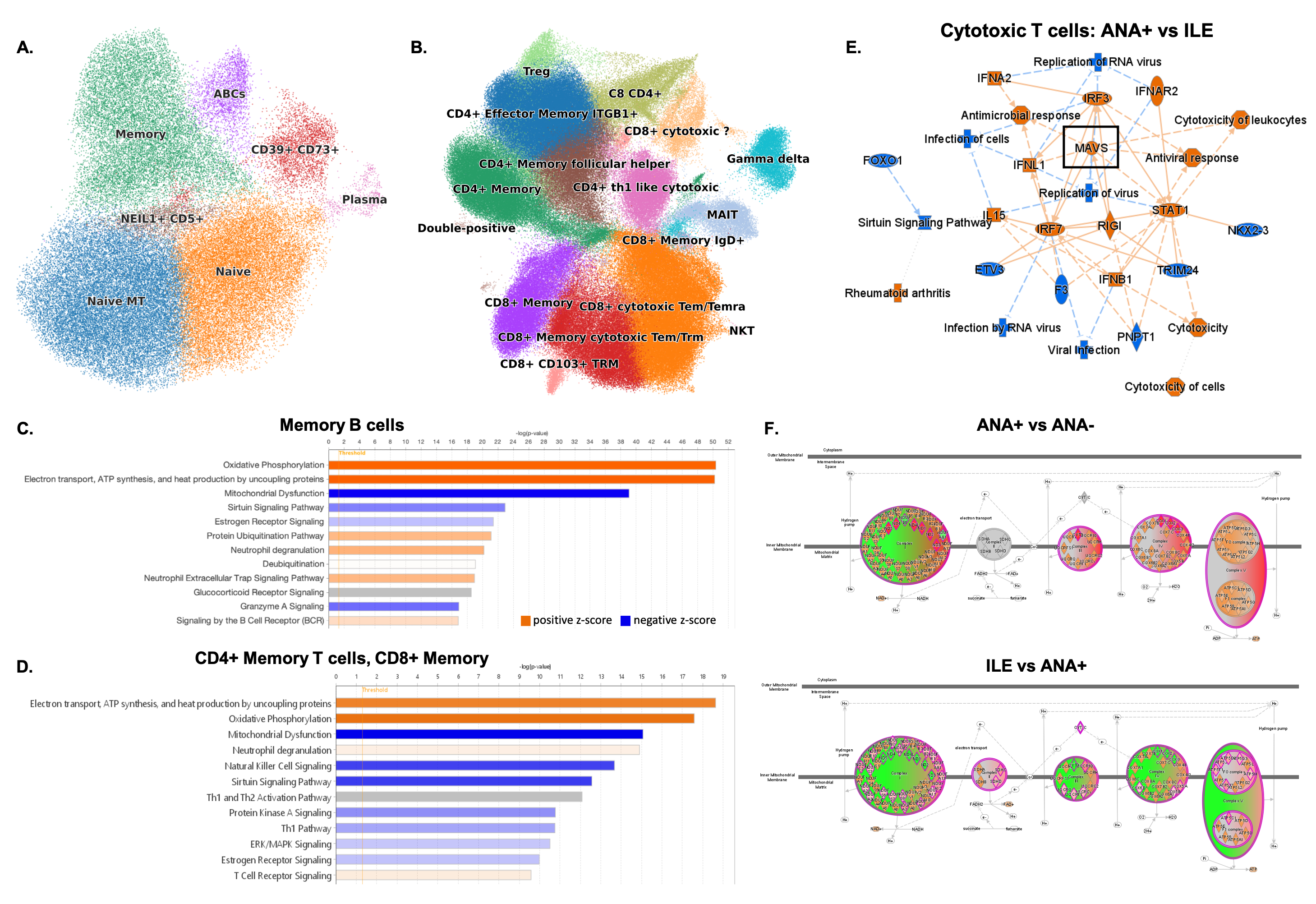
Results: Comparison of serum protein levels found the largest number of differences between ANA+ and ILE, with 478 significant proteins (Friedman non-parametric, padj < =0.05); more modest differences were noted between ANA-/ANA+ (62 proteins) and ILE/SLE (10 proteins). TAX1BP1, an autophagy receptor, appears to be the most variable protein between disease transition with covarying neighborhood analysis showing a correlation between TAX1BP1 and B cells and CD4+ memory T cell populations (Fig.1A). Pathway analysis of the most variable proteins indicated that the diversity between ANA+ and ILE is mostly driven by apoptosis, TNF-, MAPK-, NFkB signaling and interferon (IFN) activation (Fig.1B-D). Principal component analysis highlighted heterogeneity among ANA+ individuals, with one subset similar to ANA- controls, while another clustered with disease groups (Fig.1E). This second group demonstrated a disease-like signature distinguished by 900 proteins largely involved IFIH1-mediated IFN induction, nuclei-acid sensing, necrosis, and apoptosis (Fig.1F,G). Interestingly, these individuals had lower serum levels of MAVS (Fig.1H). While RIGI appears to not be involved, the data suggests activation of apoptosis and autophagy-related pathways in ILE individuals, suggesting substantial immune dysregulation (Fig.1G). Autophagy related proteins are decreased in ILE and SLE (Fig.1I). Differences in proteins involved in oxidative phosphorylation were noted in ANA+ and ILE. Correlation analysis with transcriptomic data indicates involvement of CD27+ Memory B/T cell populations (Fig.2A-D); MAVS upregulation appears to be associated with cytotoxic T cells (Fig.2E). Alterations in oxidative phosphorylation pathway differed between ANA+ from ILE in the complexes involved and suggest disrupted ATP synthesis (Fig.2F).
Conclusion: Observed signatures appear to be associated with issues with restoring cellular homeostasis and imbalance in antiviral immune response in transition between ANA+ and ILE. These abnormalities may define important events in the trajectory and stages of preclinical autoimmunity disease.
To cite this abstract in AMA style:
Bylinska A, Smith M, Lu R, Jones B, Guthridge C, Marlin M, Wright C, Macwana S, DeJager W, Beel M, Lessard C, Arriens C, Merrill J, James J, Guthridge J. Identifying Predictive Serum Soluble Mediators Signatures Specific to ANA+ at Risk of SLE Individuals with Next Generation Proteomics [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identifying-predictive-serum-soluble-mediators-signatures-specific-to-ana-at-risk-of-sle-individuals-with-next-generation-proteomics/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-predictive-serum-soluble-mediators-signatures-specific-to-ana-at-risk-of-sle-individuals-with-next-generation-proteomics/