Session Information
Date: Monday, November 13, 2023
Title: Abstracts: Pediatric Rheumatology – Clinical II: Connective Tissue Disease
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a chronic, multisystem, autoimmune disease. Childhood-onset SLE (cSLE) is characterised by a higher frequency of nervous system involvement, known as neuropsychiatric SLE (NPSLE). Biomarkers for NPSLE are lacking, posing challenges for clinical diagnosis and management. The existence of neuropsychiatric manifestations in patients without a definite NPSLE diagnosis, as well as normal conventional magnetic resonance imaging (MRI) findings in patients with NPSLE, allude to suboptimal diagnostic protocols. To identify potential biomarkers of NPSLE, we aimed to develop a deep learning model capable of identifying structural brain image features of patients with cSLE.
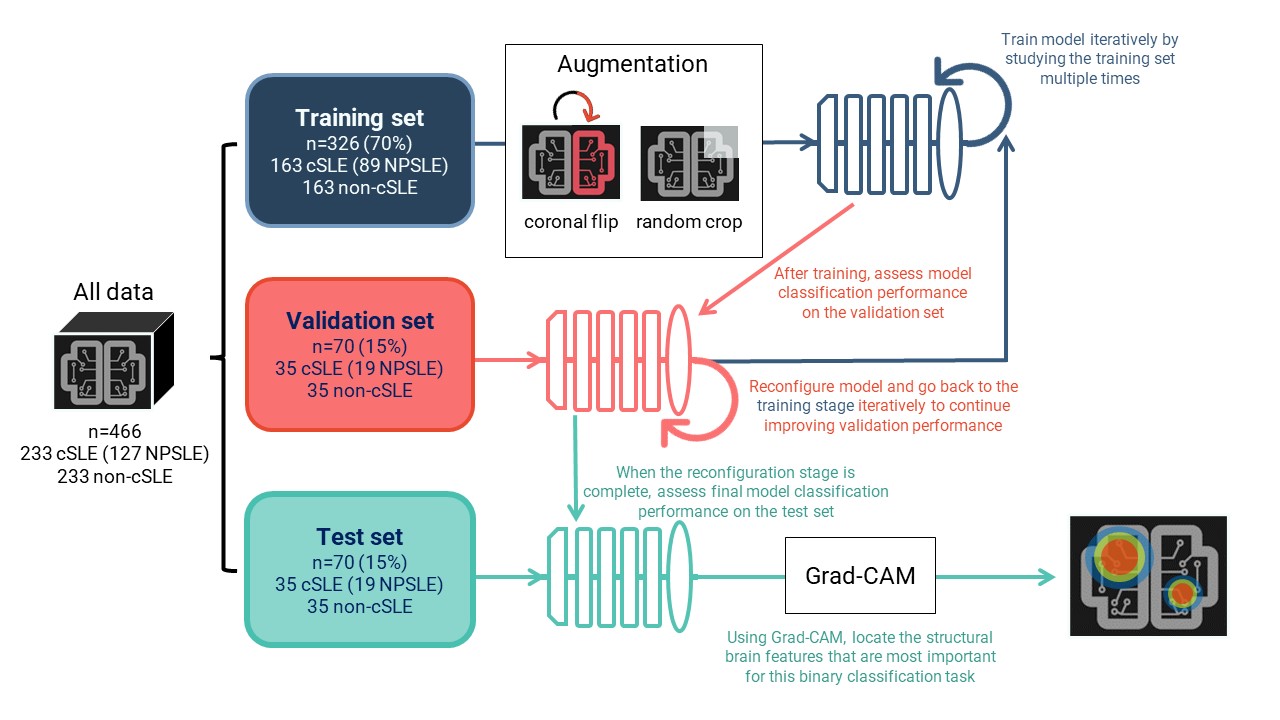
Methods: This retrospective study involved a binary classification task between cSLE and non-cSLE patients. Searching all clinically-obtained brain MRIs for cSLE patients from 2000-2021, we retrieved 233 T1-weighted images of sufficient quality for analysis. We then matched these images by age and sex to an existing set of T1-weighted images (n=233) from non-cSLE patients with clinical MRIs (primarily for indication of headache; normal conventional read), obtained from 2000-2020. A chart review was conducted to extract clinical data for cSLE patients, including history of NPSLE diagnosis and SLEDAI scores. Deep learning models consisted of a 3D image feature extractor and a linear prediction layer. The data was split into 70% for training, 15% for validation, and 15% for testing. Learning was performed on the training set. We used classification performance, measured via the ROC AUC score, on the validation set to find the best model configuration. Using the test set and the best model configuration, we assessed final model performance (Fig. 1). Interpretability analyses allowed for the identification of brain image features that were most informative for this binary classification task.
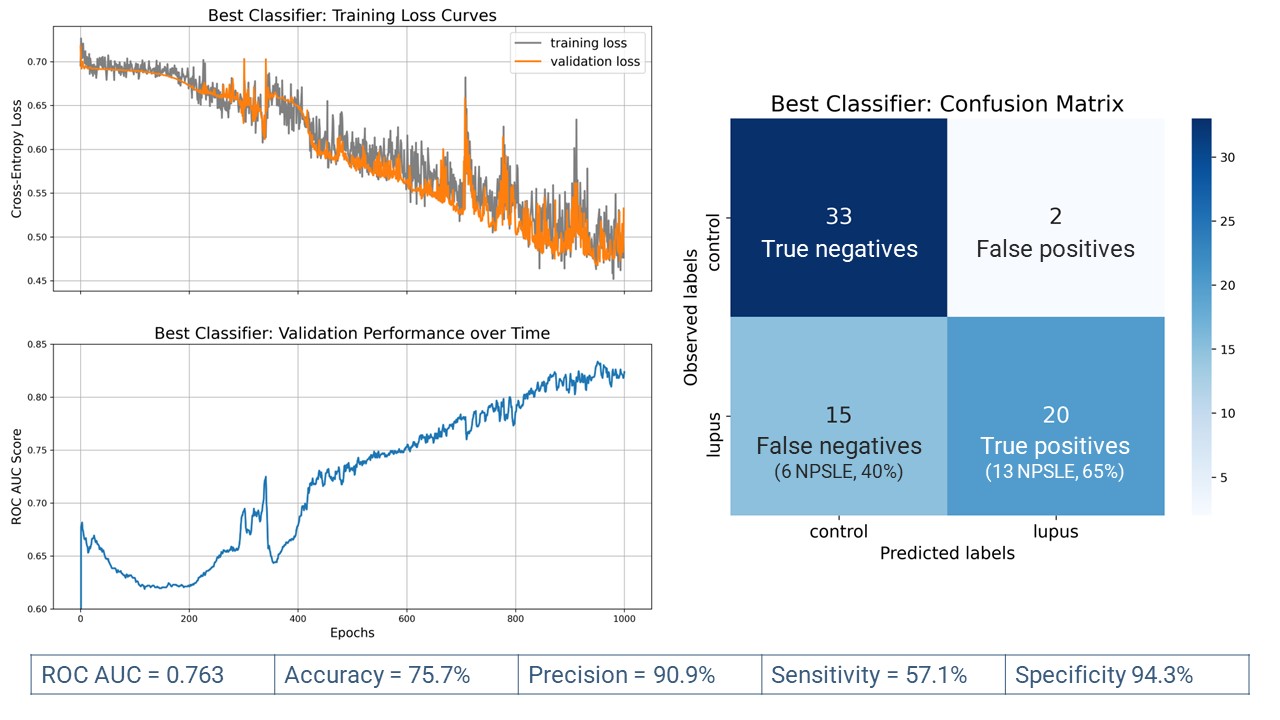
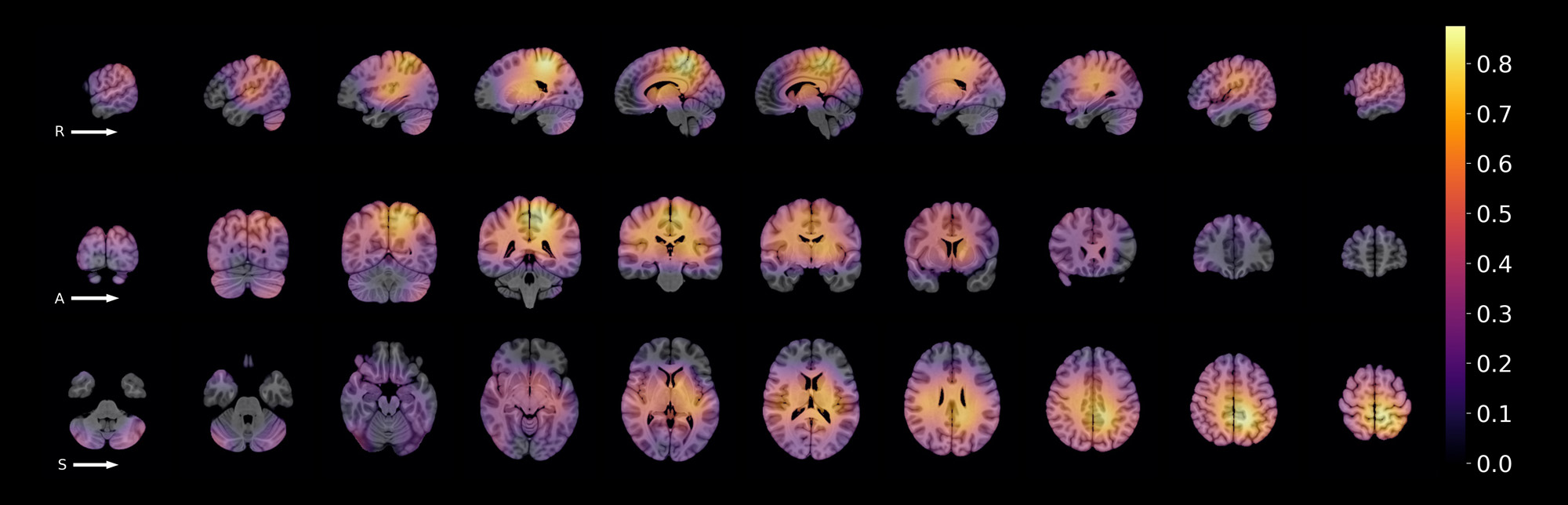
Results: Of all 233 cSLE images (133 patients), 127 (55%) were obtained from 57 patients with NPSLE. Most cSLE patients had mild/moderate disease activity at the time of scan (SLEDAI < = 10, n=153, 69.96%). When distinguishing cSLE vs non-cSLE, we achieved acceptable final classification performance with ROC AUC score = 0.763. From the 70 test samples (35 cSLE, 35 control), 20/35 of cSLE were correctly classified (true positives) (Fig. 2). The proportion of NPSLE samples in the true positives was 65%, and that in the false negatives, 40%. The difference between these proportions was not significant (z-score=1.47, p=0.07). The most important brain image features for this classification task were found in intra-ventricular, peri-ventricular, and left medial parietal regions.
Conclusion: The resulting model was effective in capturing structural brain MRI features distinguishing cSLE, with and without NPSLE, from non-cSLE patients. Our findings suggest the presence of regional brain abnormalities in cSLE patients without an NPSLE diagnosis. This model may serve as a novel diagnostic tool prototype, and as a hypothesis generating tool in future NPSLE biomarker research. Further study is needed to characterise brain image features for each type of NP manifestation observed in cSLE.
To cite this abstract in AMA style:
arciniegas s, Wagner M, Vidarsson L, Ertl-Wagner B, El Tal T, Jeyanathan A, Ng L, Hiraki L, Levy D, Ibrahim G, Knight A. Identifying Potential Neuroimaging Biomarkers of Neuropsychiatric Lupus in Children Using Deep Learning [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/identifying-potential-neuroimaging-biomarkers-of-neuropsychiatric-lupus-in-children-using-deep-learning/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identifying-potential-neuroimaging-biomarkers-of-neuropsychiatric-lupus-in-children-using-deep-learning/