Session Information
Date: Monday, November 18, 2024
Title: Abstracts: Systemic Sclerosis & Related Disorders – Clinical II
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Systemic sclerosis (SSc) is molecularly heterogeneous and distinct subtypes of patients have been identified based on gene expression in skin. Although treatments have improved, clinical trials are still confounded by significant variability that may affect their interpretation and impact. Molecular signatures based on the gene expression in skin can divide samples into four reproducible molecular intrinsic subsets (inflammatory, fibroproliferative, limited, and normal-like). The purpose of this study was to develop a stratification method using easily accessible blood samples, based on methods we pioneered in skin, that can be used to identify the patients most likely to improve with a given treatment. We tested the hypothesis that molecular heterogeneity among gene expression in skin and blood will be concordant, which can be further aid in the precision medicine and improve our ability to predict treatment responses in SSc.
Methods: We analyzed PBC samples from 68 patients enrolled in the ASSET clinical trial by RNA-seq. Samples passing QC were prepared for ribodepleted RNA-seq using the Kapa RNA HyperPrep with RiboErase + Globin protocol with 100ng RNA as input. Completed libraries were sequenced on a NextSeq2000. Data were processed using standard methods. A binary classification model was trained for each subtype using the top genes in the corresponding ranked list of differentially expressed genes for each subset.
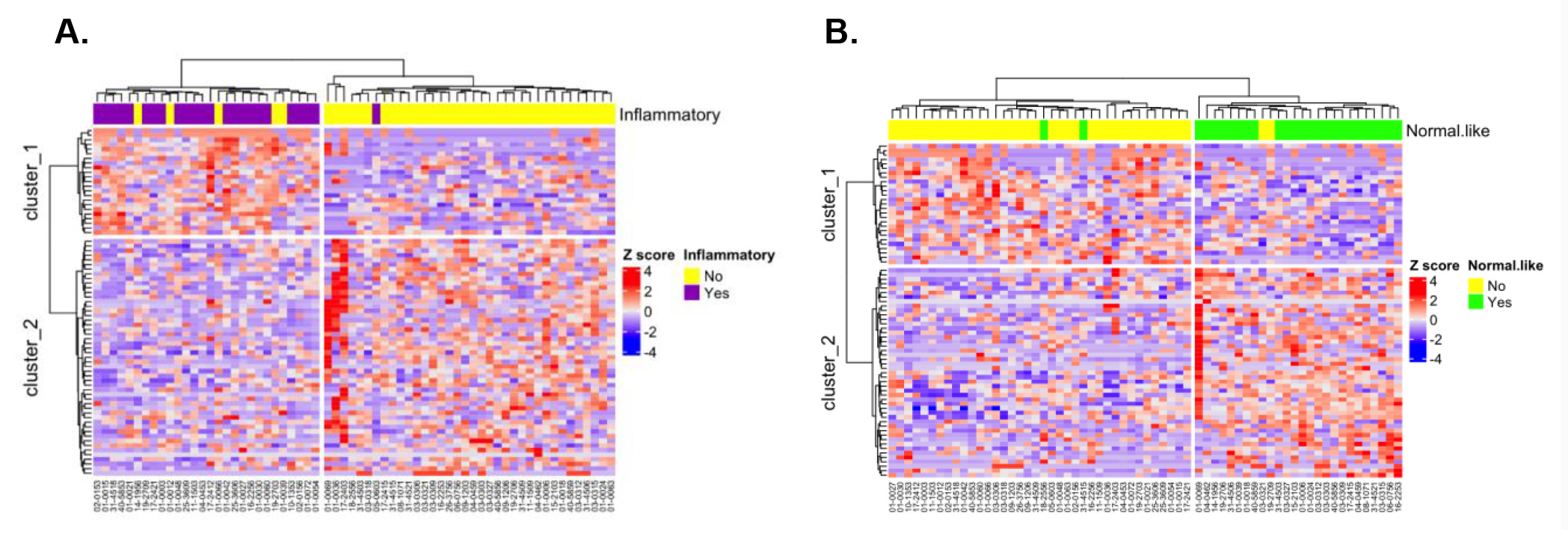
Results: RNA-seq data was generated for 68 baseline PBC samples. We focused on the 64 patients in the ASSET study who had both skin and PBC RNA-Seq data available to train a supervised learning model. The true labels used in this study were subtypes assigned to the paired skin biopsy, which was taken at the same time as the blood draw. Genes expressed in PBCs were ranked according to their correlation with each SSc intrinsic subtype predicted on their paired skin samples. A binary classification model was trained for each subtype using the top genes in the corresponding ranked list. Then, different integration methods were used to generate the final PBMC SSc intrinsic subtype prediction. We were able to achieve average accuracy around 85% using 70 genes for each SSc intrinsic subtype (Fig. 1A). By integrating results from the binary Support Vector Machine models using Max Difference method, the overall accuracy across the three subtypes was 87.5%. We further analyzed accuracy by subtype and find 83 – 88% accuracy for each individual subtype (Inflammatory: 0.83; Normal-like: 0.85; Fibroproliferative: 0.88; Fig 1B). These results confirmed our hypothesis that genes expressed in PBCs can be predictive of the SSc intrinsic subtypes originally found in the skin.
Conclusion: Our analyses using paired skin and PBC samples in untreated early SSc patients from the ASSET clinical trial show that we can predict the intrinsic molecular subsets from PBMC samples and should be validated in additional samples, including those on background immunosuppressive therapies. This provides optimism that we can use this more easily accessible tissue for subset prediction in clinical trials.
To cite this abstract in AMA style:
Gong Z, Sullivan T, Wood T, Fox D, Khanna D, Whitfield M. Identification of Systemic Sclerosis Intrinsic Subtypes in the ASSET Clinical Trial Using PBC Gene Expression [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identification-of-systemic-sclerosis-intrinsic-subtypes-in-the-asset-clinical-trial-using-pbc-gene-expression/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-systemic-sclerosis-intrinsic-subtypes-in-the-asset-clinical-trial-using-pbc-gene-expression/