Session Information
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Rheumatoid arthritis (RA) is an autoimmune disease characterized by immune infiltration of the synovial tissue, resulting in joint damage. In RA, synovia exhibit distinct cellular compositions (CTAP) (1) and variable responses to biologics. We hypothesize that RA synovia comprises distinct cellular niches, and these niches underlie synovial tissue heterogeneity and drive differential treatment response. To this end, we applied spatial transcriptomics to build a spatial atlas of RA synovia
Methods: Spatial transcriptomic profiling
We profiled RA synovial tissue samples (n=10) using CosMx to 1) define synovial cell states and 2) identify cellular regions. Next, we validated novel cellular regions identified in this dataset using Xenium on a separate RA cohort (n=7)
Cell segmentation and niche annotation
We developed a segmentation pipeline to group transcripts into cells and classified each cell into cell types, followed by a label transfer method to define fine-grained cell states. We used a graph-diffusion-based method to identify transcriptionally similar neighborhoods.
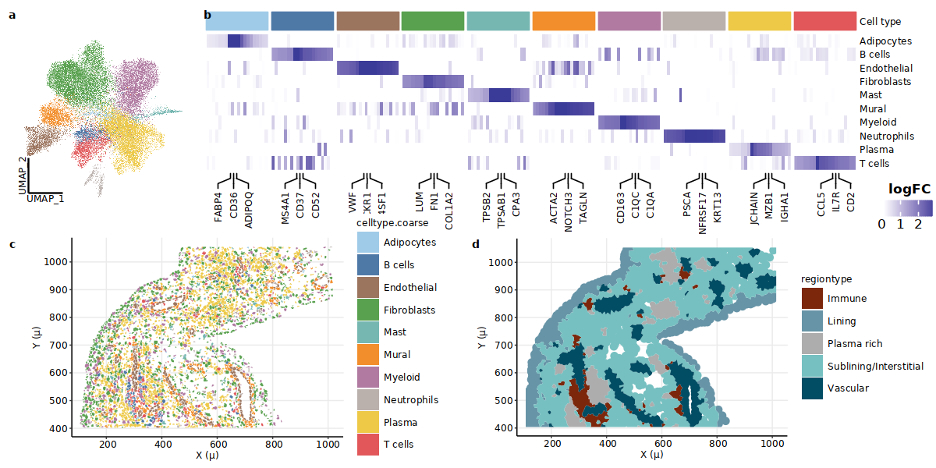
Results: Cellular niches in RA synovia. By applying spatial transcriptomic analysis followed by cellular niche analysis, we identified five spatially distinct regions:(a) lining enriched in lining fibroblasts and MERTK+ HBEGF+ fibroblasts, (b) sublining (sublining fibroblasts and myeloid cells), (c) vascular regions (endothelial and mural cells), (d) plasma cell-rich regions (plasma cells and fibroblasts), and (e) lymphoid aggregates (Fig. 1c-e).
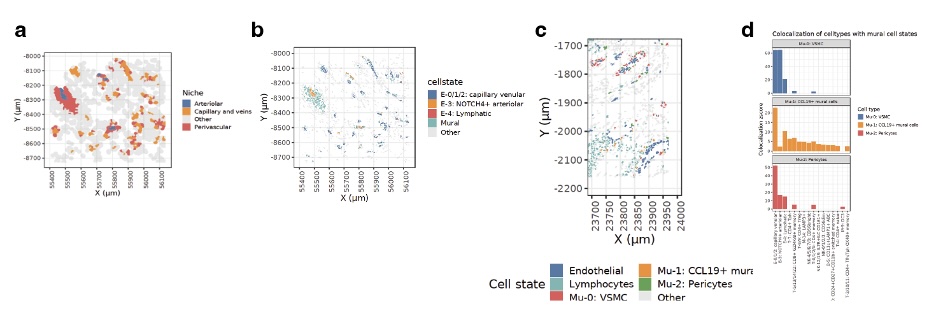
Identification of CCL19+ mural cells. Analysis of the perivascular niches revealed 3 distinct mural cell types: vascular smooth muscle cells (VSMC), pericytes, and CCL19-expressing CCL19+ mural cells (Fig. 2a). We mapped these cells in situ and identified CCL19+ mural cells are expanded near immune-infiltrated sections of synovial endothelia (Fig 2c). Colocalization analysis revealed that CCL19+ mural cells are spatially associated with T peripheral helper (Tph) cells, age-associated B cells (ABCs), and LAMP3+ DCs (Fig. 2d).
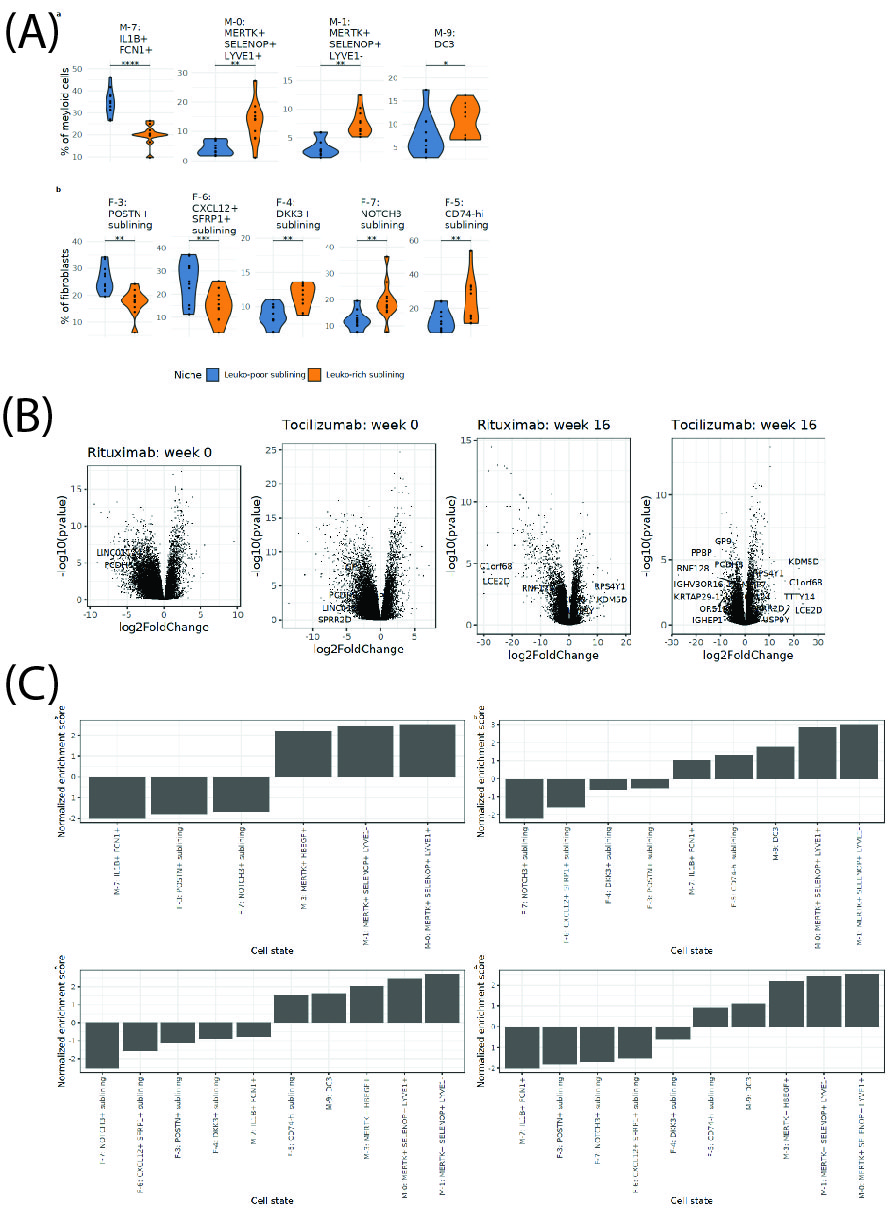
Cellular niches associated with treatment response. We defined two types of sublining niches: leukocyte-rich and leukocyte-poor. Leuko-rich niches are enriched in inflammatory DC3s, MERTK+ SELENOP+ macrophages, DDK3+ fibroblasts, NOTCH3+ fibroblasts, and CD74-hi fibroblasts, while leuko-poor subregions are enriched in CXCL12+ SFRP1+ and POSTN+ fibroblasts, and inflammatory IL1B+ FCN1+ macrophages (Fig. 3). In an independent clinical dataset of bulk RNA-seq from the R4RA trial, we found the gene signatures for leukocyte-rich enriched populations consistently overexpressed in synovial samples from responders (vs non-responders), both before and after treatment and for both Rituximab or Tocilizumab (Fig. 4 and5).
Conclusion: We established the first spatial atlas of RA synovia at a single-cell resolution. Identifying cellular niches enriched in responders and non-responders to biologics will improve our understanding of cellular and molecular pathways driving treatment outcomes in RA.
(B) Gene expression in responsders vs non-responders. Volcano plot of differential expression genes (responders vs non-responders) for Rituximab and Tocilizumab at two time points for two different drugs.
(C) Normalized enrichment scores (NES) of the transcriptional profile of cell states in non-responders. (a) NES for cell state transcriptional profiles in patients treated with Rituximab at week 0. (b) NES for cell state transcriptional profiles in patients treated with Tocilizumab at week 0. (c) NES for cell state transcriptional profiles in patients treated with Rituximab at week 16. (d) NES for cell state transcriptional profiles in patients treated with Tocilizumab at week 16.
To cite this abstract in AMA style:
Korsunsky i, Madhu R, Wei K, Bhamidipati K, Tran M, Presti S, Jonsson A, Gravallese E, Brenner M, Raychaudhuri S, Seifert J, Pitzalis C, Zhang F, Moreland L, Holers V, Clayman M, Lewis M, Gao C, Zuscik M, Autoimmune and Immune-Mediated Disease A. Identification of Spatial Determinants of Treatment Response in Rheumatoid Arthritis Synovia [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identification-of-spatial-determinants-of-treatment-response-in-rheumatoid-arthritis-synovia/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-spatial-determinants-of-treatment-response-in-rheumatoid-arthritis-synovia/