Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Immunohistochemistry is the gold standard for antibody staining in microscopy. However, a lack of multiplexing limits its utility in the use of discovery. Here we demonstrate a serial Immunohistochemistry (sIHC) staining workflow and analysis pipeline allowing for multiplexing of 18 antibodies on a single section.
Methods: 29 FFPE block were included for preliminary analyses from the 127 clinically indicated kidney biopsies recruited as part of the Lupus nephritis (LN) Accelerating Medicines Partnership (AMP) project).
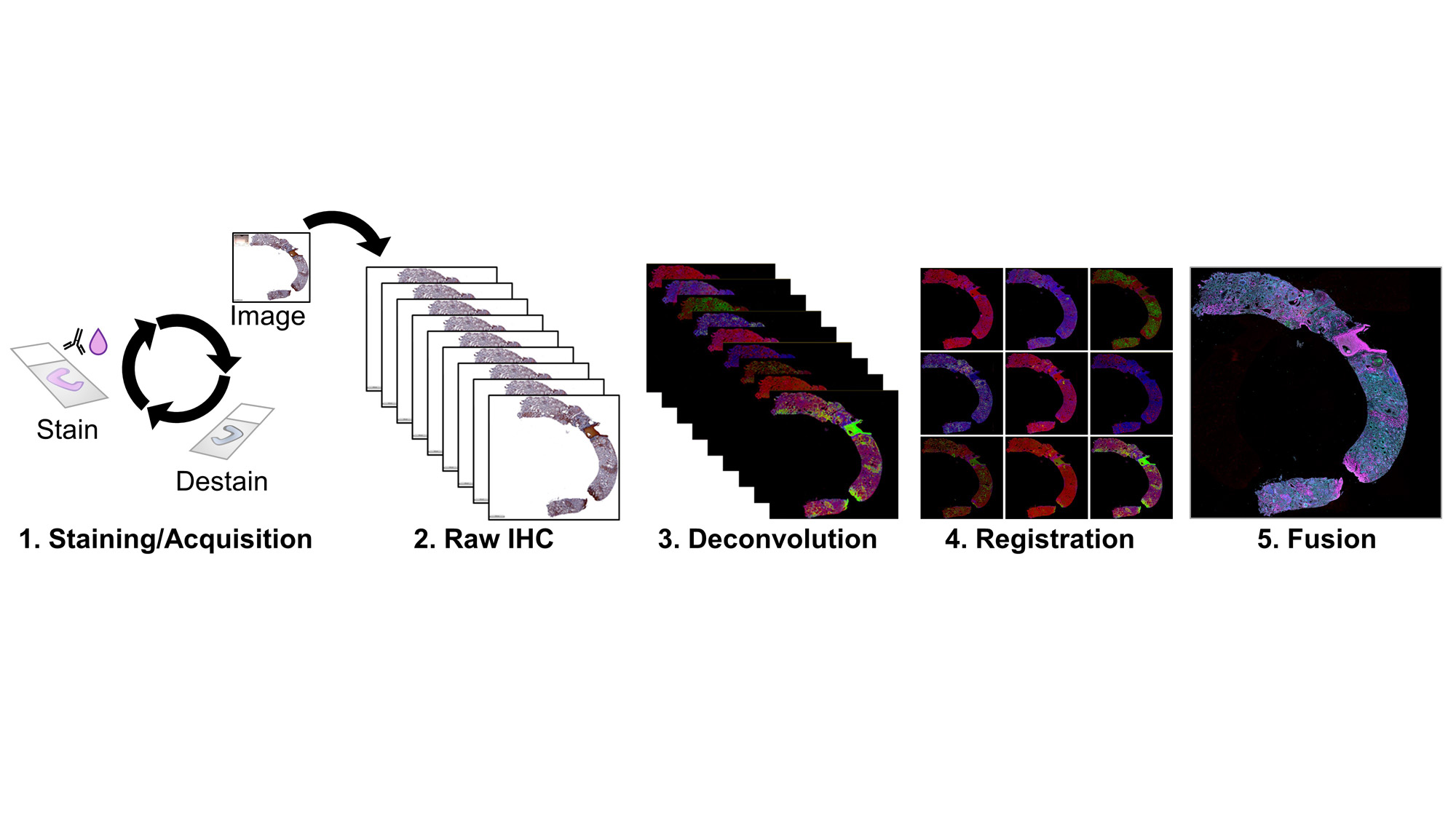
Staining: 10mm sections from LN kidney biopsies in FFPE were deparaffinized, rehydrated, and antigen retrieved with boiling citrate. Slides were blocked for peroxidases, washed, and incubated with primary antibody, secondary HRP reagents, AEC-Red Chromogen, and Hematoxylin. Images were acquired at 40X magnification. Slides were decolored in 90% Ethanol and stripped in an antibody elution buffer, then cycled for all markers.
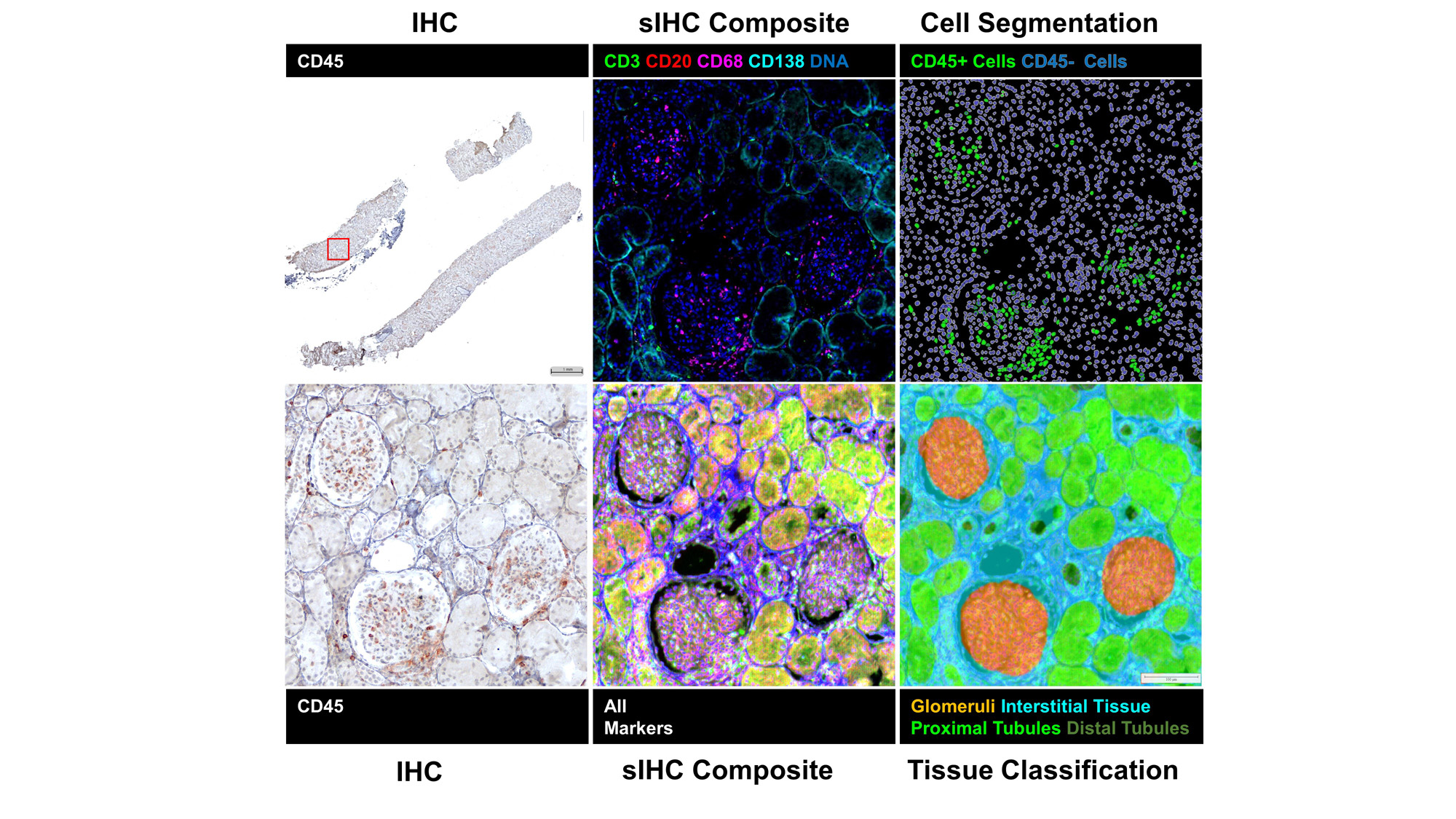
Image Stacks: Image files were imported into HALO-AI v3.5 (Indica Labs) for deconvolution, pseudo-coloring, co-registration, and alignment. Registered images were fused to create a single composite image with all 18 markers and a representative DNA and counterstain channel. Tissue regions were identified by a DenseNet V2 AI tissue classifier trained on an original co-registered IHC image and individual cell objects were identified by nuclear detection and segmented by cell distance boundaries.
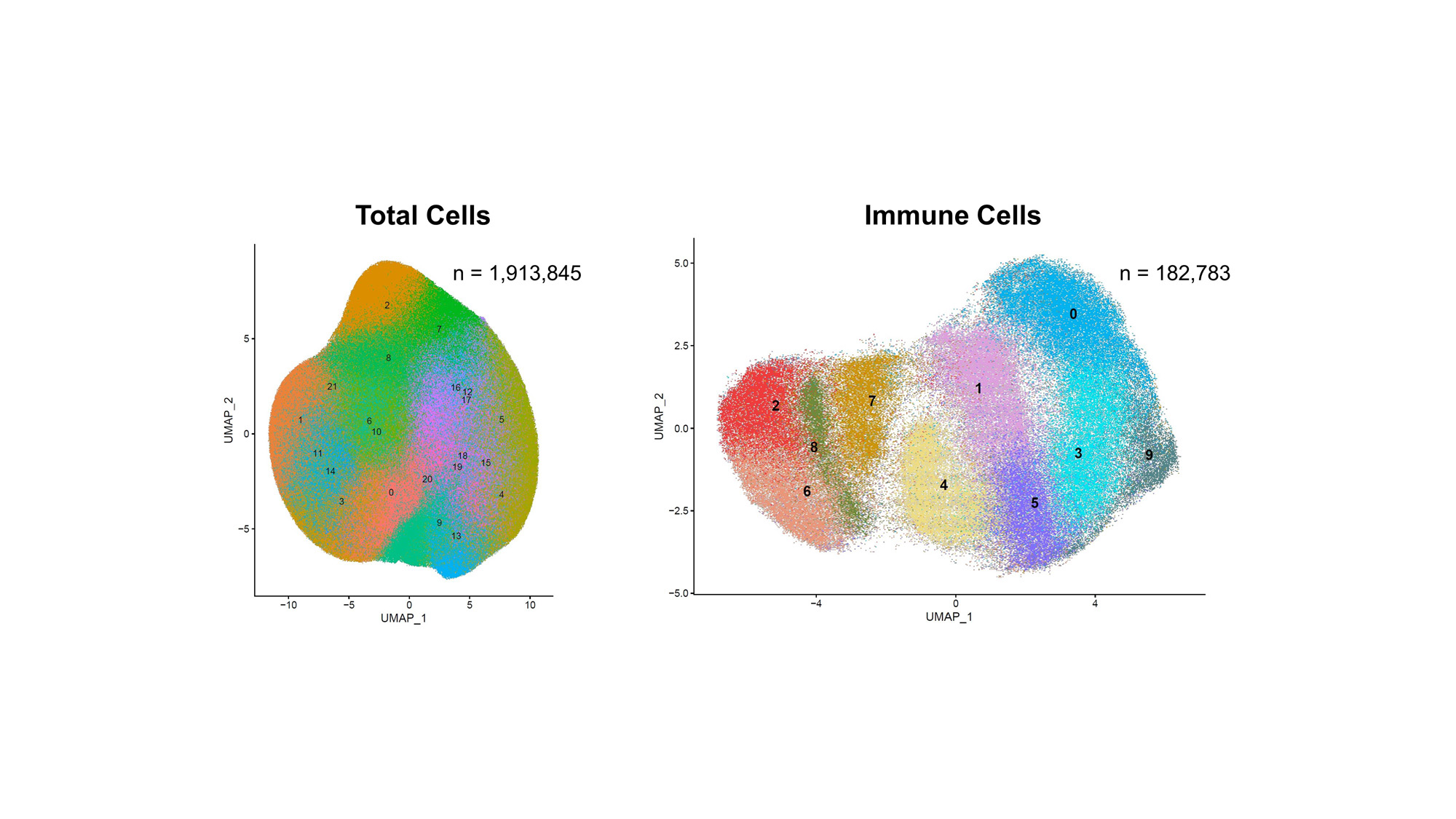
Analysis: Single-cell marker expression datasets were analyzed in R. Cells located on the edge of the tissue or in regions classified as wrinkles were removed to reduce potential artifacts. Marker expression values underwent CLR-normalization and standardization within each sample and then combined and harmonized by Harmony to further minimize batch effect. PCA and UMAP were used for dimensional reduction, and KNN and SNN algorithms were applied to identify cell clusters. Clusters with CD45 expression levels > 2 standard deviations were selected as candidate immune cell clusters. Cell clusters with high keratin marker expression were removed to eliminate renal tubular epithelial cells. Remaining clusters were then re-clustered via KNN and SNN to identify distinct immune cell types with the help of heatmaps and feature plots.
Results: The co-registration and fusion of images produced image stacks in which all 20 markers could be visualized within the same image. Tissue classification and single-cell segmentation allowed for clear identification of regions of the tissue and the individual cells that constituted each region. Single-cell analyses identified 1,913,845 cells and 182,783 CD45+ cells which were classified into 10 distinct immune cell phenotypes.
Conclusion: Here we demonstrate imaging and analysis of 20 markers using sIHC for the first time, while producing a large, spatially informed, clinically-relevant dataset in 29 LN Kidney Biopsies.sIHC can be successfully employed to perform multiplexed whole slide analysis harnessing both the subcellular resolution (brightfield) and the reliability of IHC.
To cite this abstract in AMA style:
Marlin C, Celia A, Furie R, James J, Fava A, Guthridge J, Rosenberg A, Stephens T, Wright C, Hodgin J, Izmirly P, Belmont H, Anolik J, Petri M, Buyon J, Kamen D, Putterman C, The A, Lee C, Yang X. Identification of Lupus Nephritis Kidney Immune Populations via Hi-Resolution 20-Plex Immunohistochemistry Single-Cell Spatial Analyses [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/identification-of-lupus-nephritis-kidney-immune-populations-via-hi-resolution-20-plex-immunohistochemistry-single-cell-spatial-analyses/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-lupus-nephritis-kidney-immune-populations-via-hi-resolution-20-plex-immunohistochemistry-single-cell-spatial-analyses/