Session Information
Date: Wednesday, November 15, 2023
Title: Abstracts: Vasculitis – Non-ANCA-Associated & Related Disorders III: Innovation
Session Type: Abstract Session
Session Time: 11:00AM-12:30PM
Background/Purpose: The availability of diagnostic laboratory tests and specific biomarkers of disease activity for giant cell arteritis (GCA) remains an area of unmet need. The purpose of this study was to utilize a high-throughput screening array to identify plasma proteins that 1) differentiate patients with GCA from controls; and 2) associate with disease activity in GCA.
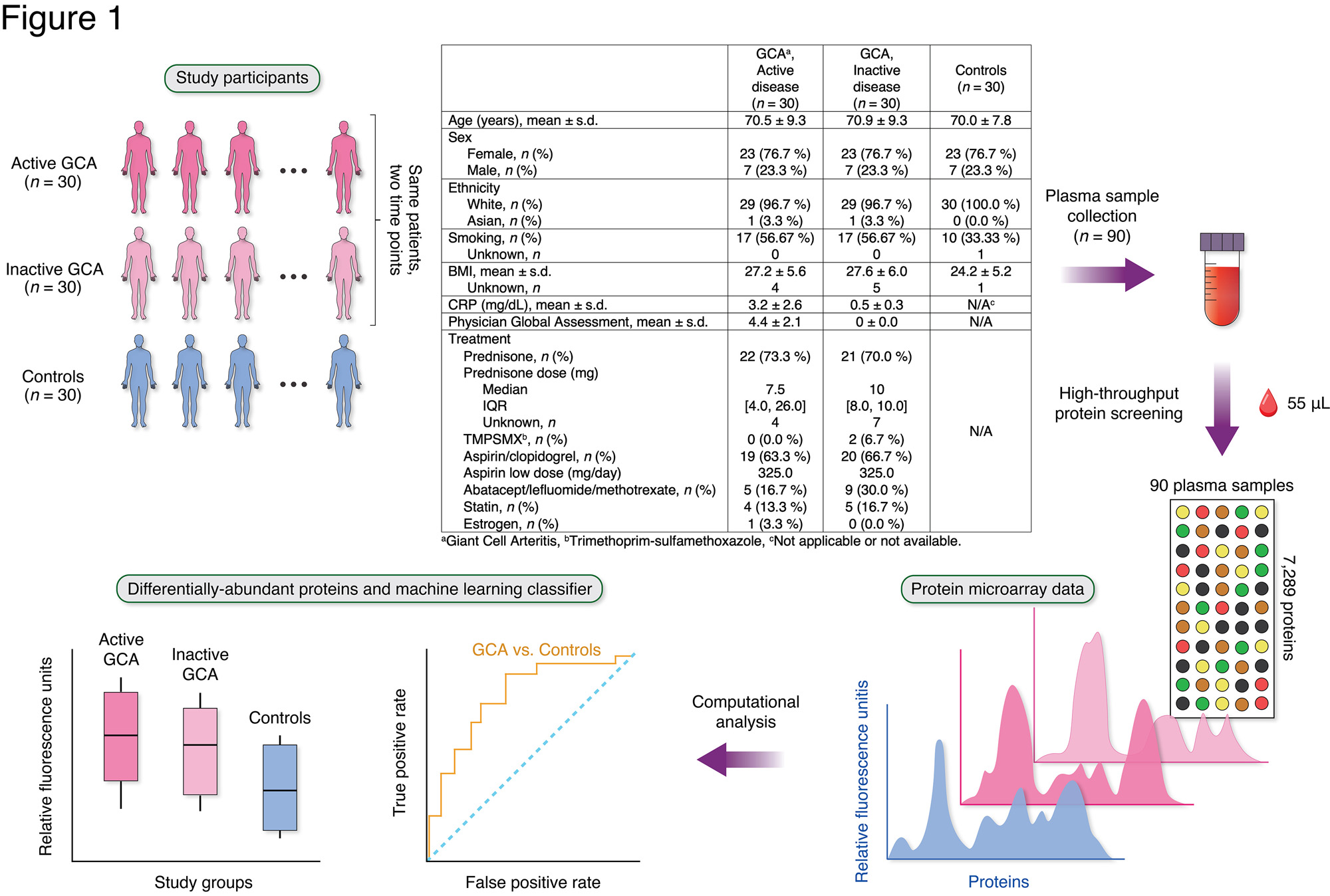
Methods: This study included patients with GCA (n = 30) from a multi-institutional prospective longitudinal cohort study and 30 age-/sex-/race-matched healthy controls (Figure 1). Most patients with GCA were taking glucocorticoids at the time of sample collection (Figure 1). Plasma samples were collected from patients with GCA at two separate visits: 1) during active disease; and 2) during inactive disease. An aptamer-based, multiplex microarray platform (SomaScan® Assay, SomaLogic) measured semi-quantitative abundances of 7,289 proteins in relative fluorescence units (RFUs). Linear regression models identified differentially-abundant proteins between patients with GCA (at active or inactive disease state) compared with controls while adjusting for potential confounders (P < 0.01). Gene Ontology (GO) Biological Processes identified enriched functional categories of the differentially-abundant proteins. Proteins associated with disease activity (Physician Global Assessment, PGA) in patients with active GCA were also identified. A random forest model was trained on plasma proteomes to develop a classifier that distinguishes GCA (active or inactive state) from controls.
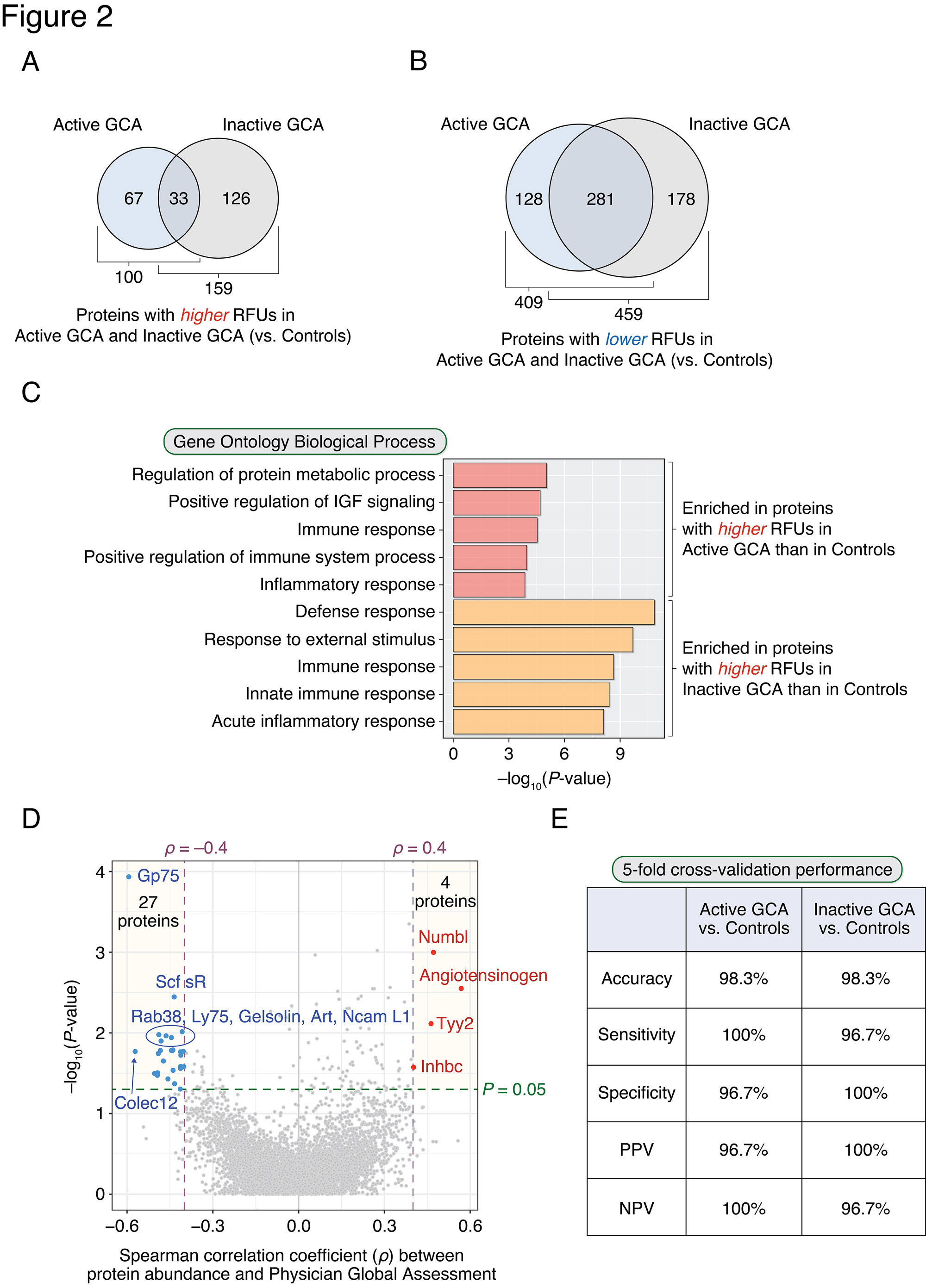
Results: 509 and 618 differentially-abundant proteins were identified between active GCA compared with healthy controls (Figure 2A) and inactive GCA compared with controls (Figure 2B), respectively. Among these, 100 and 159 proteins had significantly higher abundances in active GCA and inactive GCA, respectively. The observed enriched biological processes are shown in Figure 2C: ‘Regulation of protein metabolic process’ and immune-related processes were the most highly enriched functions of plasma proteins found higher in active GCA. Interestingly, processes involving apoptosis, nucleotide metabolism, and EGFR, ErbB, and neurotrophin signaling were also significantly enriched. A paired analysis between active and inactive visits found 219 differentially abundant proteins. In addition, 31 proteins were found to be associated with disease activity (PGA) in patients with active GCA (Figure 2D). A random forest classifier correctly predicted active GCA vs. controls with an accuracy of 98.3% (sensitivity: 100%; specificity: 96.7%) in 5-fold cross-validation (Figure 2E). Similarly, in the case of inactive GCA vs. controls, a random forest classifier distinguished these two phenotypes at 98.3% accuracy (sensitivity: 96.7%; specificity: 100%).
Conclusion: Plasma proteome profiling in two different disease states of GCA produced highly accurate classification for distinguishing active and inactive disease states from controls. These results demonstrate the strong potential of integrating plasma proteomes with machine learning for future approaches to identify GCA. Future studies should include validating these findings in a larger independent cohort.
To cite this abstract in AMA style:
cunningham k, Sung J, Hur B, GUPTA V, Koster M, Weyand C, Cuthbertson D, Khalidi N, Koening C, Langford C, McAlear C, Monach P, Moreland L, Pagnoux C, Rhee R, Seo P, Merkel P, Warrington K. Identification of Giant Cell Arteritis Using Plasma Proteome Profiles Integrated with Machine Learning [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/identification-of-giant-cell-arteritis-using-plasma-proteome-profiles-integrated-with-machine-learning/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-giant-cell-arteritis-using-plasma-proteome-profiles-integrated-with-machine-learning/