Session Information
Date: Saturday, November 6, 2021
Title: Spondyloarthritis Including PsA – Basic Science Poster (0046–0068)
Session Type: Poster Session A
Session Time: 8:30AM-10:30AM
Background/Purpose: Psoriatic arthritis (PsA) is an inflammatory arthritis that currently lacks diagnostic biomarkers. We hypothesized that there are differences in the protein intensities in the synovial fluid (SF) of patients with osteoarthritis (OA), rheumatoid arthritis (RA) and PsA. We aimed to identify markers for PsA by proteomic analysis of knee joint SF. Our objectives were to identify proteins and pathways differentially expressed between:
1. OA and the inflammatory arthritides (PsA and RA).
2. PsA and RA.
Methods: Mass spectrometry was used to identify the SF proteome from 10 OA, 10 PsA, and 10 RA patients. All patients satisfied the classification criteria for their disease. First, we used student’s t-test for differential intensity analysis (p-value < 0.05; fold change > 1.5) to identify potential protein biomarkers of each disease. Next, we performed protein-protein interaction (PPI) network and pathway enrichment analysis to obtain biological insights on systems biology of these diseases.
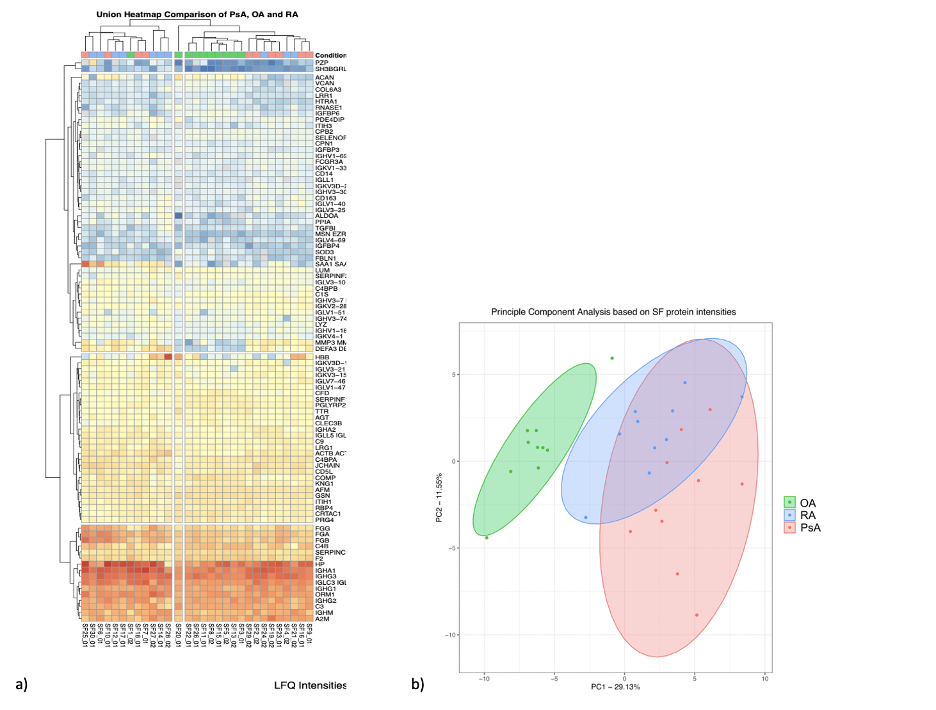
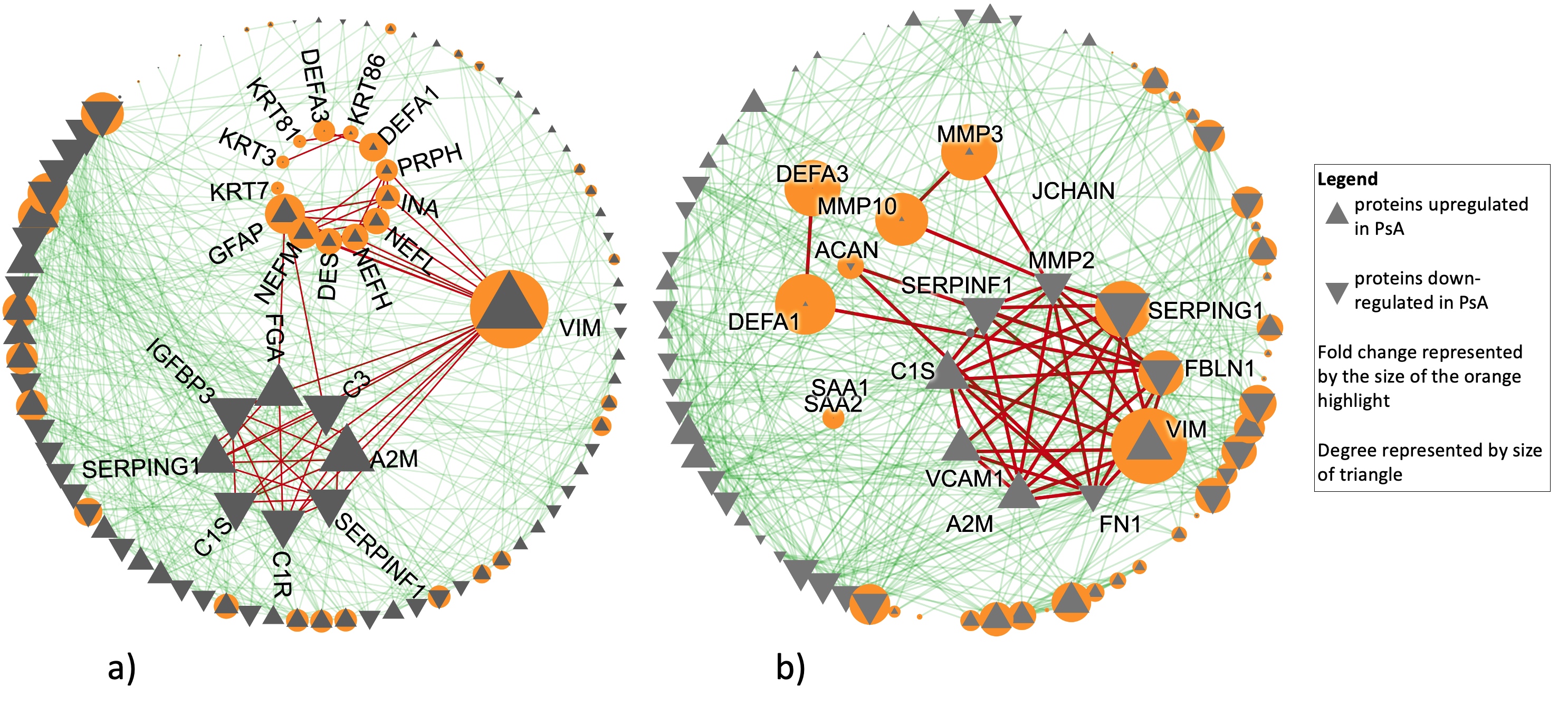
Results: Aim 1: 4 proteins were present and 45 were absent in OA SF compared to PsA and RA. Among those present in all conditions, we detected 56 and 66 proteins with differential intensities in OA compared to PsA and RA, respectively. Hierarchical clustering and principal component analysis confirmed the power of these proteins in distinguishing OA from the inflammatory arthritides (Fig 1). In both comparisons, enriched pathways were involved in inflammation and immune response. Additionally, PPI network analysis of both lists revealed connected subnetworks of proteins with high fold change enriched in immune-system and extracellular matrix (Fig 2a,b). In both comparisons, VIM showed the highest fold change and was central in connecting other high fold change proteins.
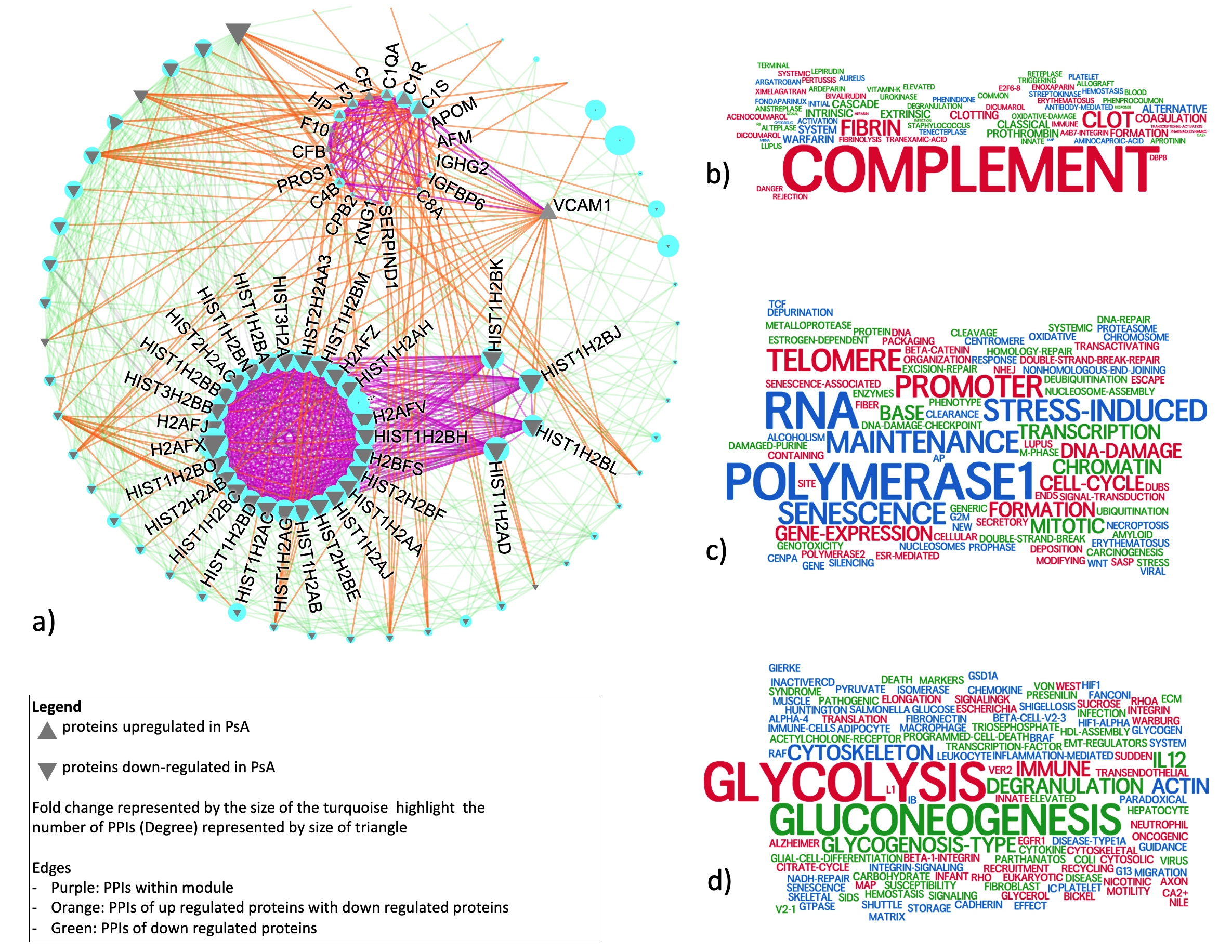
Aim 2: 22 proteins with differential intensities between PsA and RA were identified. In addition to immune system pathways, these proteins were enriched in cell metabolism pathways, such as glycolysis which has been previously suggested to distinguish PsA and RA. PPI analysis identified three densely connected network modules (Fig 3a). All upregulated proteins in PsA formed one dense module enriched in complement and clotting pathways (Fig 3b). We identified one highly densely connected subnetwork enriched in transcription-related pathways (Fig 3c) which has four interactions with one upregulated protein (VCAM1). All other proteins were downregulated and have many interactions with the upregulated module. These proteins were highly enriched in glycolysis and glycogenesis, followed by immune pathways (Fig 3d). Previous research has shown interplay between glycolysis and complement pathways. Unlike in Aim 1, VIM shows small but statistically significant differences.
Conclusion: We have identified differential proteins and pathways between PsA, OA, and RA. VIM was a central protein in connecting proteins with differential intensities between OA and the inflammatory arthritides. Additionally, we identified three large network modules enriched in complement, transcription, and glycolysis pathways that may distinguish PsA from RA. Our results suggest disrupted interplay between complement and glycolysis pathways in PsA vs RA.
To cite this abstract in AMA style:
Lai J, Rahmati S, Sivakumar R, Oikonomopoulou K, Abji F, Chandran V. Identification of Biomarkers and Deregulated Pathways in Psoriatic Arthritis Through Proteomic Analysis of Synovial Fluid [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/identification-of-biomarkers-and-deregulated-pathways-in-psoriatic-arthritis-through-proteomic-analysis-of-synovial-fluid/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-biomarkers-and-deregulated-pathways-in-psoriatic-arthritis-through-proteomic-analysis-of-synovial-fluid/