Session Information
Date: Saturday, November 6, 2021
Title: Spondyloarthritis Including PsA – Basic Science Poster (0046–0068)
Session Type: Poster Session A
Session Time: 8:30AM-10:30AM
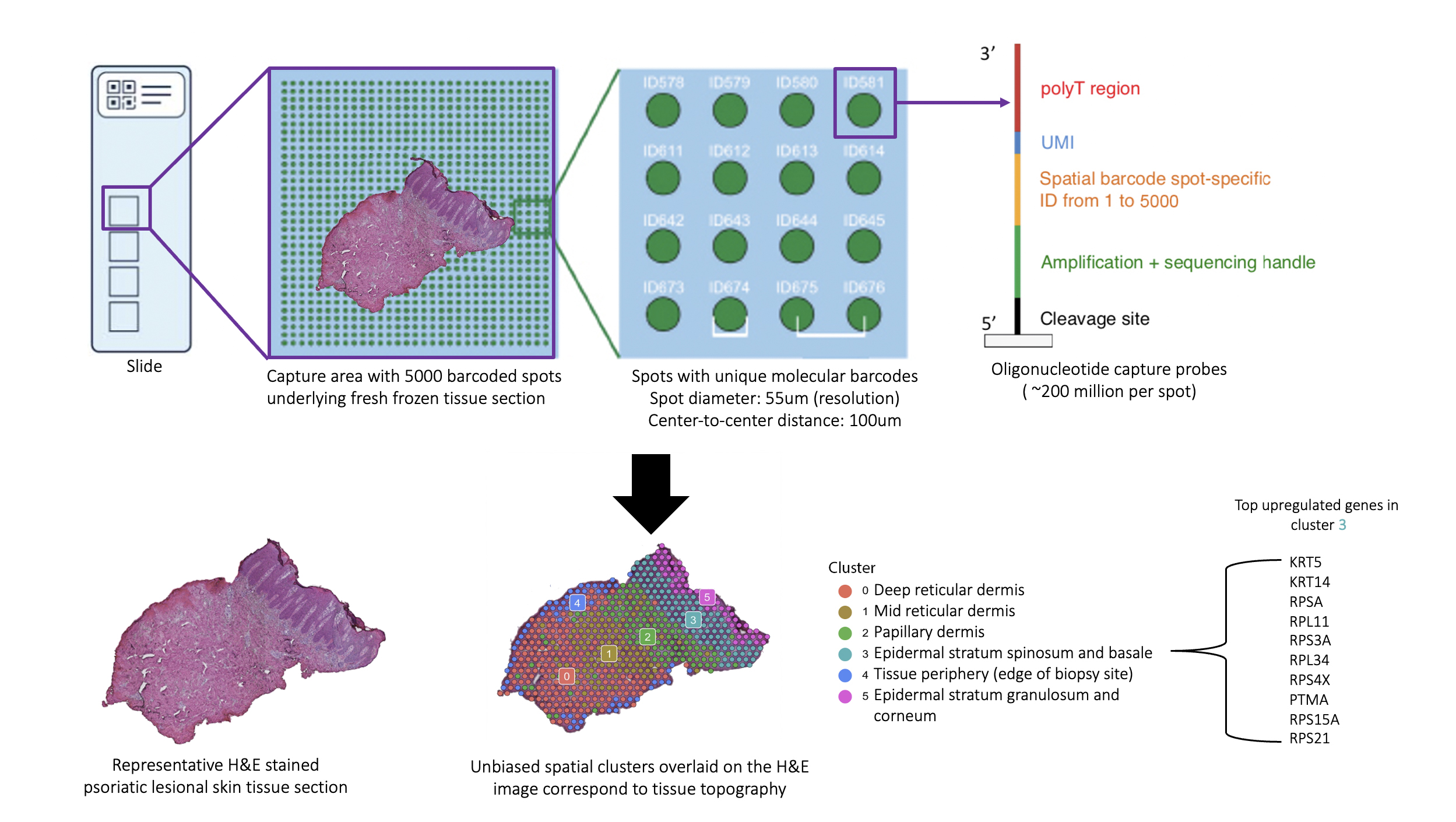
Background/Purpose: The skin is recognized as a window into the immunopathogenic mechanisms in the psoriatic arthritis (PsA) joint. This is evidenced by the fact that skin disease precedes joint involvement in ~90% of PsA patients and by the greater degree of overlap between gene expression in the synovial tissue of PsA and lesional skin in psoriasis (PsO) compared to synovium in other forms of inflammatory arthritis.1 Thus, the study of the psoriatic skin transcriptome has the potential to yield revolutionary insights into the immunopathogenesis of PsA. Spatial transcriptomics (ST) is a ground-breaking technology that allows for mRNA sequencing from histologically-intact tissue sections, facilitating precise localization of the site of gene expression (Fig. 1). A platform for ST (Visium Spatial Gene Expression Solution by 10X Genomics) has been made available; however, it remains to be optimized for human skin tissue, which is inherently challenging to use in transcriptomic studies due in part to its preponderance of RNAses.2,3 To date, there are no published studies on ST in either healthy or psoriatic human skin. Through a multidisciplinary collaboration, we have successfully optimized both healthy and psoriatic human skin tissue for use with ST.
Methods: Workflow
- Skin biopsy
- Cryopreservation
- Sectioning and staining
- Confirmation of RNA integrity
- Optimization of permeabilization
- Sequencing
- Data analysis
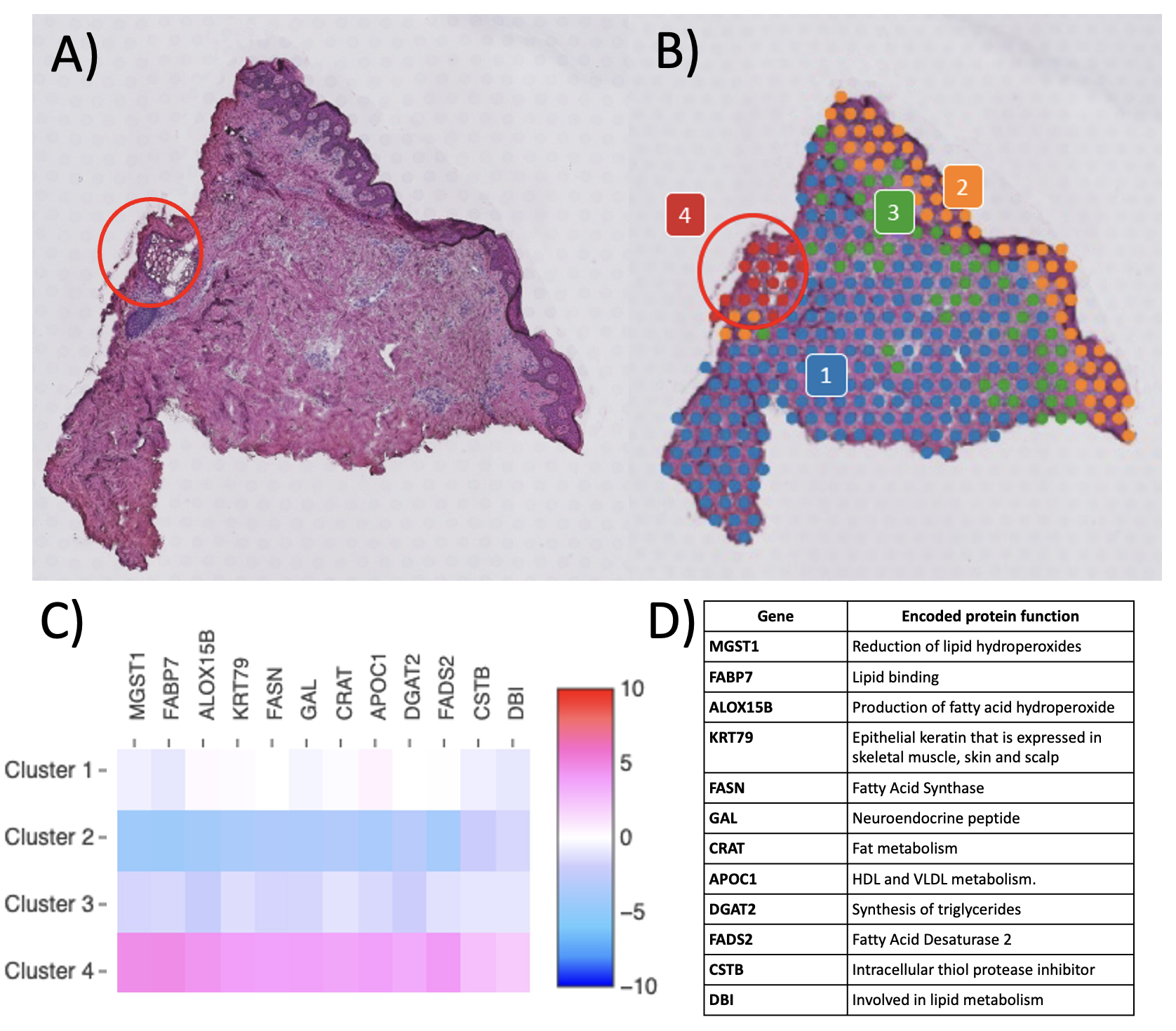
Results: To date, we have accrued samples from 3 controls, 4 PsA patients, and 6 PsO patients. All samples met the platform’s quality control (QC) metrics for spots and mapping, with reads in spots under tissue >50% (range: 63.1 to 87.1) and reads mapped confidently to exonic regions >30% (range: 80.1 to 91.5). The expected biological variation in transcriptional activity as evidenced by molecular counts (which correlate strongly with unique genes)4 across disease states and tissue regions was observed, with gene expression strikingly greater in the cell-dense epidermis and in appendageal structures than in the dermis and in psoriatic lesional skin compared to non-lesional and control skin (Fig. 2A). To detect technical variation, two contiguous sections from the same control sample were run on separate slides (Fig. 2B). Clustering and the Uniform Manifold Approximation and Projection (UMAP) plot architecture were consistent between the two replicates. Importantly, accuracy of spatial localization of gene expression and biological consistency of unbiased clustering was observed, with concordance of histopathologically annotated regions with gene-expression based clustering (Fig. 3).
Conclusion: Successful optimization of both healthy and psoriatic human skin tissue for ST was achieved, with all samples meeting the platform’s QC metrics. The expected biological variance in transcriptional activity across tissue regions and disease states was noted, the quantity, quality, and location of reads was biologically consistent, and there was no technical variation between samples. Thus, spatial profiling of gene expression in psoriatic skin through spatial transcriptomics can be performed and has the potential to offer invaluable insights into the immunopathogenesis of psoriatic disease.
To cite this abstract in AMA style:
Castillo R, Sidhu I, Yan D, Konieczny P, Haberman R, Hsieh B, Neimann A, Naik S, Scher J. Harnessing Spatially-Resolved Gene Expression to Characterize the Transcriptional Landscape of Psoriatic Skin [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/harnessing-spatially-resolved-gene-expression-to-characterize-the-transcriptional-landscape-of-psoriatic-skin/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/harnessing-spatially-resolved-gene-expression-to-characterize-the-transcriptional-landscape-of-psoriatic-skin/