Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic sclerosis (SSc) displays significant heterogeneity. Those with limited cutaneous systemic sclerosis (lcSSc) experience significant systemic organ involvement similar to those with diffuse cutaneous systemic sclerosis (dcSSc). LcSSc with anti-centromere antibody (ACA) is associated with lower rates of overall clinical organ manifestations and better survival compared to those without ACA. Here we explore whether skin biopsy gene expression might account for these differences.
Methods: Bulk RNA sequencing was undertaken on uninvolved skin biopsies from 14 lcSSc patients with ACA (ACA+), 14 lcSSc without ACA (ACA-) (3 anti-topoisomerase (ATA); 1 anti-polymerase III (ARA); 3 ANA positive, ENA negative; 3 anti-Pm-Scl; 1 ANA negative; 2 anti-ribonucleoprotein (RNP); 1 anti-fibrillarin (U3-RNP)) and 5 healthy controls (HC). Differential gene expression was undertaken using R and DESeq2 with false discovery rate < 0.05, analysed with uncorrected p values ≤ 0.001. Gene functional enrichment analysis was undertaken with gProfiler (p< 0.05).
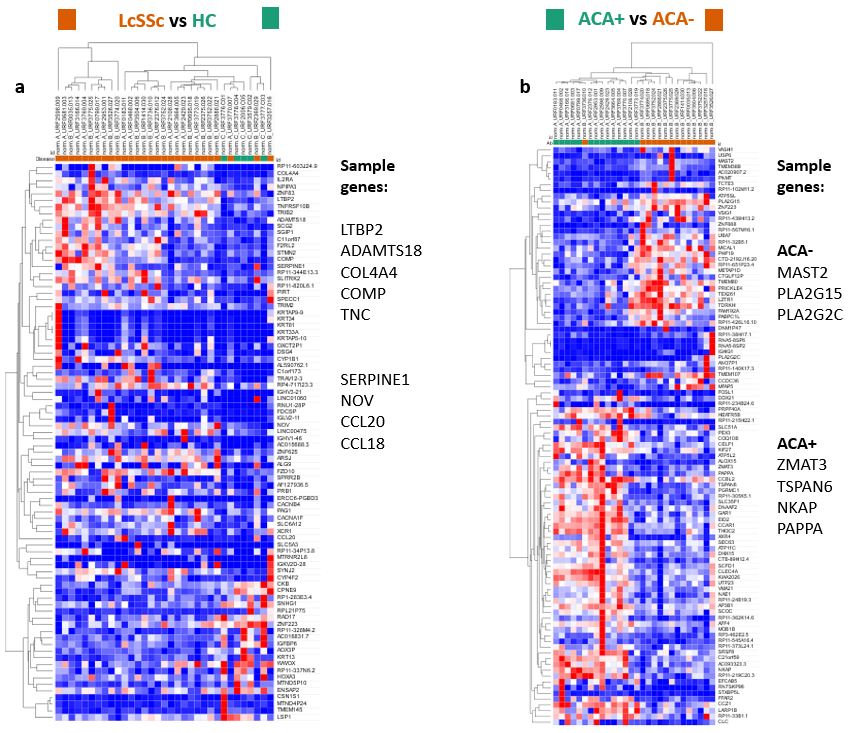
Results: Unsupervised cluster analysis of the whole cohort showed, as previously reported, considerable overlap between HC and lcSSc. However, within the lcSSc samples there was significant separation of ACA+ and ACA- samples (Figure 1). Therefore, we compared these directly with HC and delineated differences in gene expression dependent upon ACA status (Figure 2).
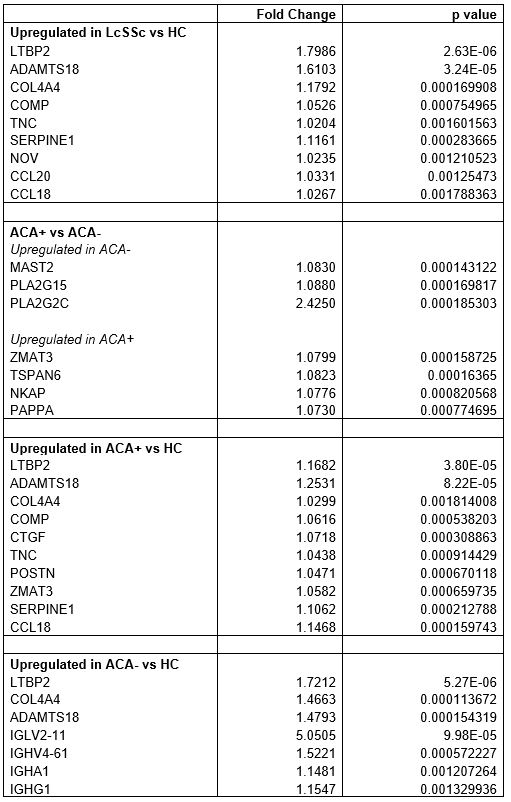
Compared to HC, lcSSc patients showed upregulation in genes implicated in SSc pathogenesis (Table 1). LTBP2, ADAMTS18, COL4A4, COMP and TNC are involved in extracellular matrix organisation, cell adhesion and fibroblast to myofibroblast differentiation. SERPINE1, NOV, CCL18 and CCL20 are involved in monocyte chemotaxis and CCL18 derived macrophage polarisation towards a fibrogenic M2 phenotype has been implicated as a marker of lung fibrosis.
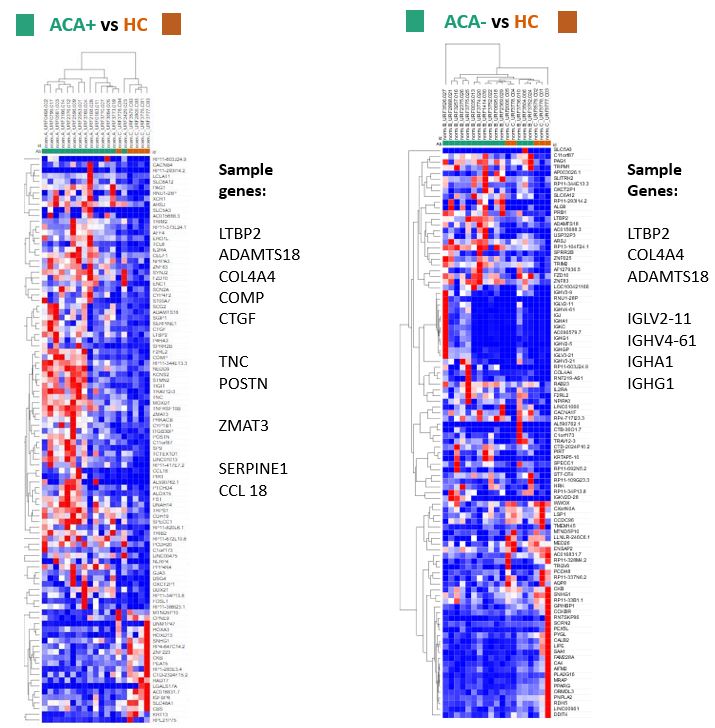
Differences between ACA+ biopsies and ACA- were apparent comparing within the lcSSc samples and when separate comparisons were made for each antibody subset and HC. ACA- compared to ACA+ showed overexpression of MAST2, a regulator of SERPINE1 expression, and PLA2G15 and PLA2G2C, with serum phospholipase A2 a mediator of vascular inflammation. ACA+ samples relative to ACA-, overexpressed ZMAT3, a positive regulator of p53; TSPAN6, expressed in vascular injury and NKAP and PAPPA, both involved in the activation of NF-kappaB.
ACA+ against HC additionally showed overexpression of CTGF and POSTN associated with pathogenesis of cardiovascular disease and TGFβ/Smad signalling. ACA- against HC particularly enriched for pathways related to immunoglobulins and B cell receptor and complement activation (IGLV2-11, IGHV4-61, IGHA1, IGHG1).
Conclusion: We confirm key SSc-associated differences in gene expression for lcSSc that align with previous reports and pathogenic pathways seen in dcSSc. We identify differences between ACA+ and ACA- lcSSc skin biopsy gene expression that may predict different clinical outcomes in these subgroups and help to stratify patients for clinical trials and clinical care.
To cite this abstract in AMA style:
Yee P, Kanitkar M, Clark K, Ong V, Denton C. Genomic Analysis of Skin Biopsies Differentiates Major Anti-Nuclear Autoantibody Subsets in Limited Cutaneous Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/genomic-analysis-of-skin-biopsies-differentiates-major-anti-nuclear-autoantibody-subsets-in-limited-cutaneous-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genomic-analysis-of-skin-biopsies-differentiates-major-anti-nuclear-autoantibody-subsets-in-limited-cutaneous-systemic-sclerosis/