Session Information
Date: Monday, November 14, 2022
Title: Abstracts: SLE – Diagnosis, Manifestations, and Outcomes III: Genetic Factors
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a polygenic autoimmune disease whose specific causes are incompletely understood and for which there exists no single comprehensive diagnostic test. Primary immunodeficiency diseases (PID) are severe failures of the immune system caused by single-gene defects. Genes with impaired function in PID may represent a rich source of candidates for genes overexpressed in SLE. The goal of this study was to investigate whether these genes were overrepresented in SLE patient gene expression.
Methods: A comprehensive database of 453 PID genes was compiled based on previous reports published by the International Union of Immunological Societies (IUIS) committee as well as review of published autoimmune disease literature. This database was analyzed for biological function enrichment using gene set variation analysis (GSVA) and compared to known SLE risk genes and SLE patient differential gene expression (DE). PID genes were grouped using protein-protein interaction network clustering, and the resulting clusters were tested by GSVA and machine learning to determine their efficacy as focused diagnostic gene lists.
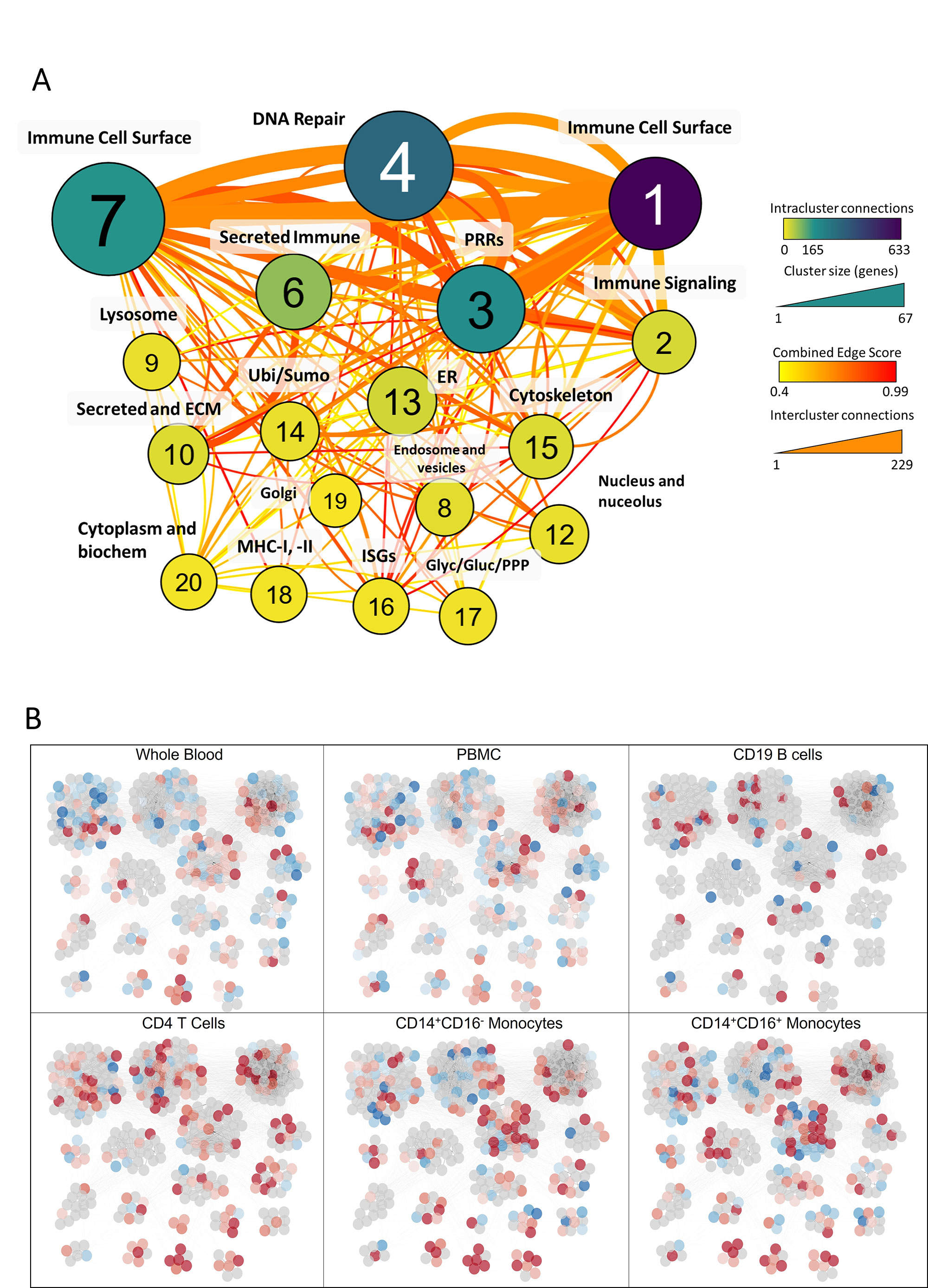
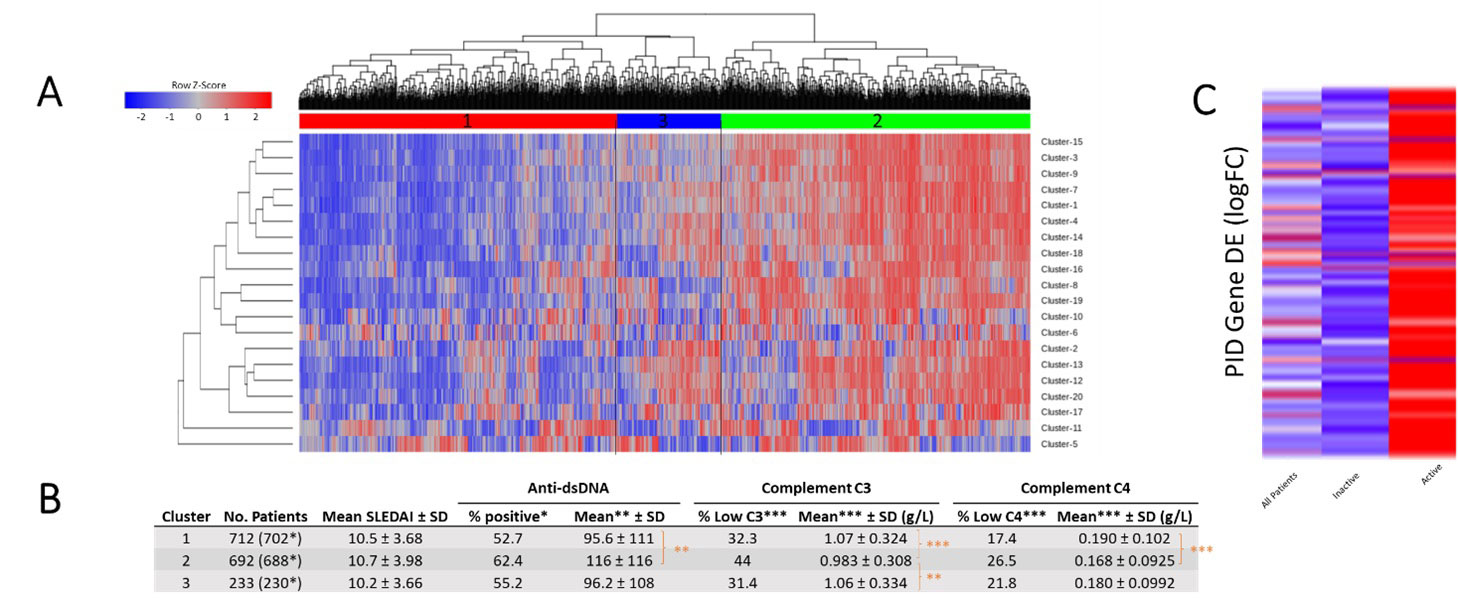
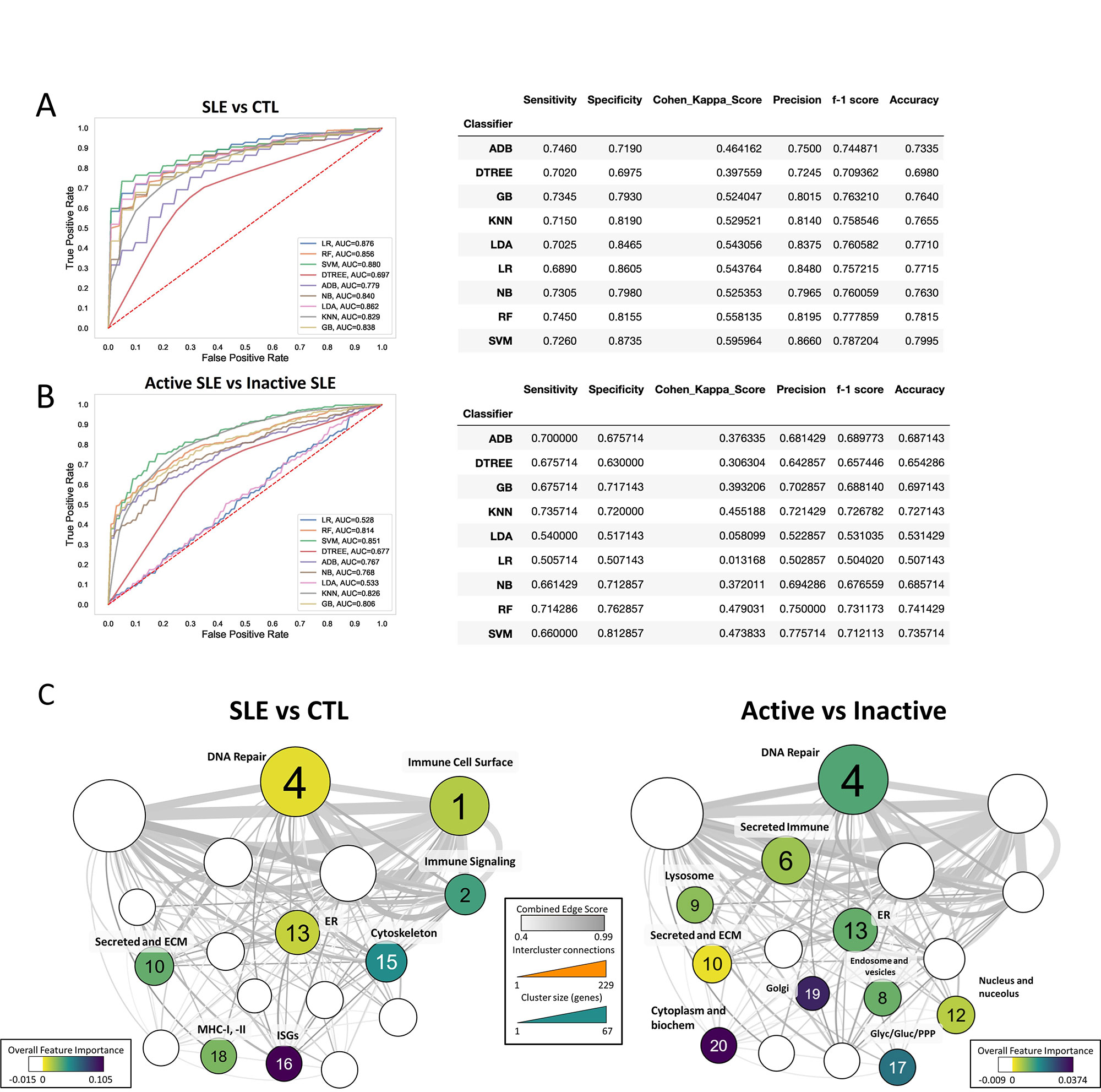
Results: Genes included in the PID Database were highly enriched for functional ontologies and pathways involved in immune cell function and specific to immune cell populations. mCODE clustering produced 16 distinct cellular and functional groups which were differentially enriched in SLE patient subpopulations (Fig 1). PID genes also showed disproportionate overlap with SNP-predicted SLE risk genes, with 119 (26.2%) of the PID genes identified as lupus risk genes. The majority of PID genes were overexpressed in SLE patients and especially in those with active disease (SLEDAI >6) (Fig 2). Finally, functional groups of PID genes defined by mCODE were employed as features for machine learning and effectively classified patients as having lupus or not or having active or inactive SLE (Fig 3). Notably, different sets of clusters were effective in each classification.
Conclusion: PID genes overlap SLE risk genes, are over-expressed in SLE and can be used in machine learning models to classify SLE. PID genes, thereby, represent a key set of genes involved in lupus pathogenesis.
To cite this abstract in AMA style:
Davis H, Labonte A, Owen K, Hubbard E, Kain J, Kegerreis B, Bachali P, Grammer A, Lipsky P. Genes Causative of Primary Immunodeficiency Are Risk Factors for and Over-expressed in Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/genes-causative-of-primary-immunodeficiency-are-risk-factors-for-and-over-expressed-in-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genes-causative-of-primary-immunodeficiency-are-risk-factors-for-and-over-expressed-in-systemic-lupus-erythematosus/