Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose: Lupus neutrophils play an important role in tissue damage (including glomerulonephritis and lupus skin involvement), as well as endothelial damage in lupus patients. Several studies have suggested altered functional capacity of lupus neutrophils compared to healthy controls, and the expansion of a neutrophil subset with a lower density (low density granulocytes or LDGs) in lupus. We characterized the DNA methylome of normal-density neutrophils and LDGs from lupus patients, and age-, sex-, and ethnicity-matched healthy controls to gain insight into chromatin architectural differences and how these differences might alter neutrophil function and induce pathogenicity in lupus.
Methods: We included 8 lupus patients and 8 healthy age-, sex-, and ethnicity-matched controls for these studies. Low density granulocytes (LDGs) and normal-density neutrophils were isolated from each lupus patient, and normal-density neutrophils were isolated from healthy controls. Neutrophils and LDGs were extracted using density gradient centrifugation and magnetic bead separation with over 95% purity in every sample. DNA was extracted and bisulfite conversion performed. A genome-wide DNA methylation study was performed using the Illumina HumanMethylation 450 BeadChip array, which includes over 485,000 methylation sites across the entire genome. The array covers 99% of RefSeq genes, including promoter regions, 5′-UTR, first exon, gene body, and 3′-UTR. 96% of all CpG islands are also covered. Statistical and bioinformatics analysis was performed to identify differentially methylated loci, gene ontologies, and pathways.
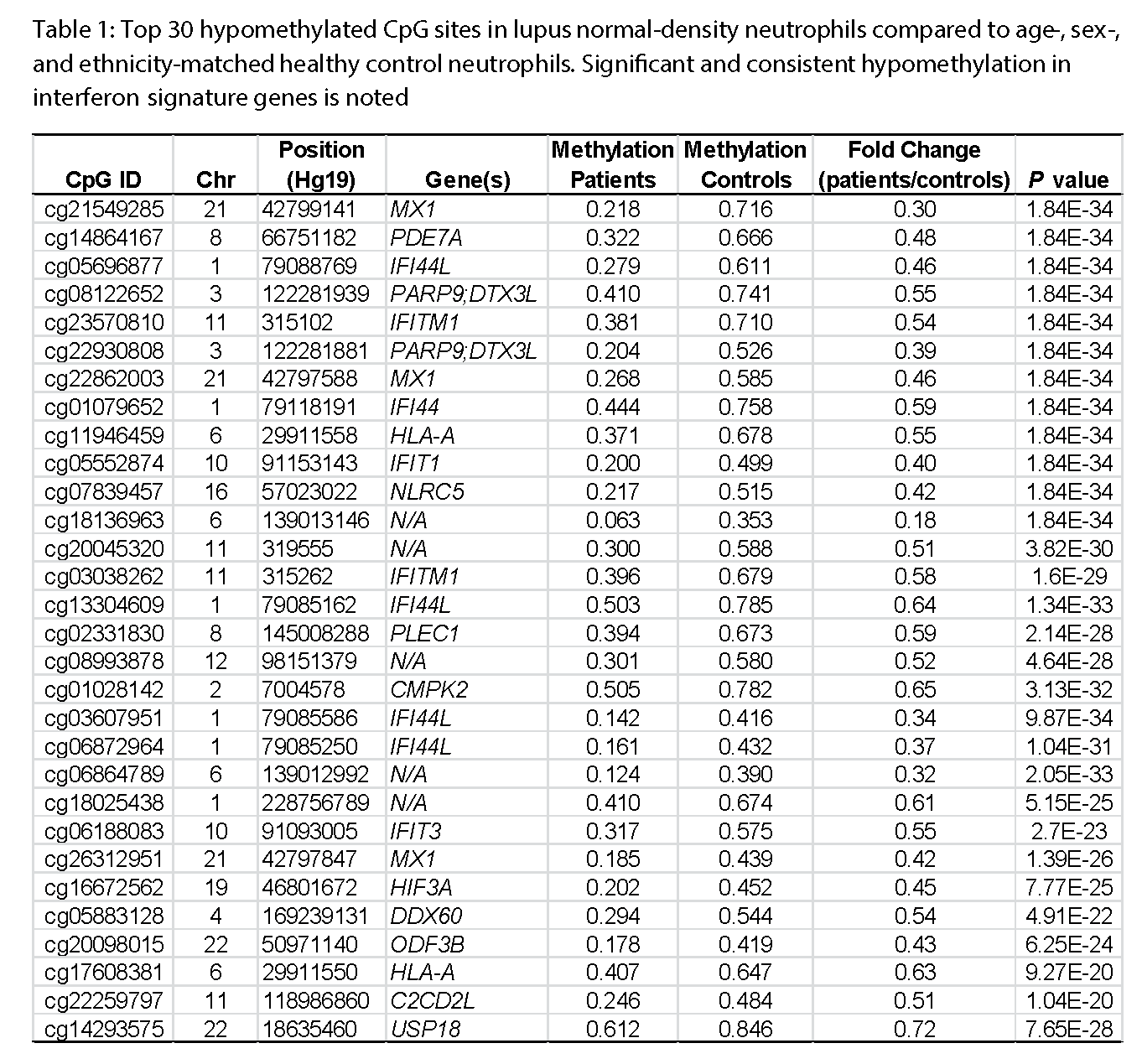
Results: We identified 619 differentially methylated CpG sites in normal-density neutrophils between lupus patients and healthy matched controls. Very interestingly, we find a robust and consistent demethylation of interferon signature genes in both normal-density neutrophils and LDGs in lupus patients. The top 30 hypomethylated CpG sites in lupus neutrophils are presented in Table 1. These data suggest that the chromatin structure in lupus granulocytes supports the expression of higher levels of type-1 interferon regulated genes, and that neutrophils directly contribute to the interferon signature in lupus. Curiously, however, our data so far suggest that LDGs and normal-density neutrophils from lupus patients are virtually identical in their chromatic architecture with no methylation differences across the genome between these two distinct granulocyte subsets.
Conclusion: We characterized the DNA methylome in lupus neutrophils for the first time and showed a pattern of robust demethylation of interferon signature genes in lupus patients supporting a pathogenic role for neutrophils in lupus.
Disclosure:
P. Coit,
None;
S. Yalavarthi,
None;
W. Zhao,
None;
M. J. Kaplan,
None;
A. H. Sawalha,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/epigenome-profiling-reveals-robust-hypomethylation-of-interferon-signature-genes-in-lupus-neutrophils/