Background/Purpose: Giant cell arteritis (GCA) is a systemic large vessel vasculitis of unknown etiology. A hallmark of GCA is the presence of granulomatous inflammation of the arterial wall. The DNA methylation status of the arterial environment in GCA patients has not been previously elucidated. This study characterizes the DNA methylation profile of formalin-fixed, paraffin-embedded (FFPE) temporal artery tissue biopsies taken from GCA patients and age, sex, and ethnicity matched controls that presented with similar symptoms, but had normal temporal artery biopsies.
Methods: Temporal artery biopsies performed at the University of Michigan from 1988-2012 were reviewed. Twelve patients with non-equivocal histological evidence for GCA and twelve age, sex, and ethnicity matched controls with normal biopsies were included in this study. All patients fulfilled the ACR classification criteria for GCA and were not taking steroids for more than 48 hours prior to biopsy. DNA was extracted from the affected portions of FFPE temporal artery tissue in GCA patients and from histologically-confirmed normal arteries in controls. Genome-wide DNA methylation status was evaluated using the Illumina Infinium HumanMethylation 450 BeadChip Array, which covers 99% of RefSeq genes and over 485,000 methylation sites across the genome. Differentially methylated loci between affected and unaffected arterial tissues were identified, and subsequent bioinformatic analysis performed.
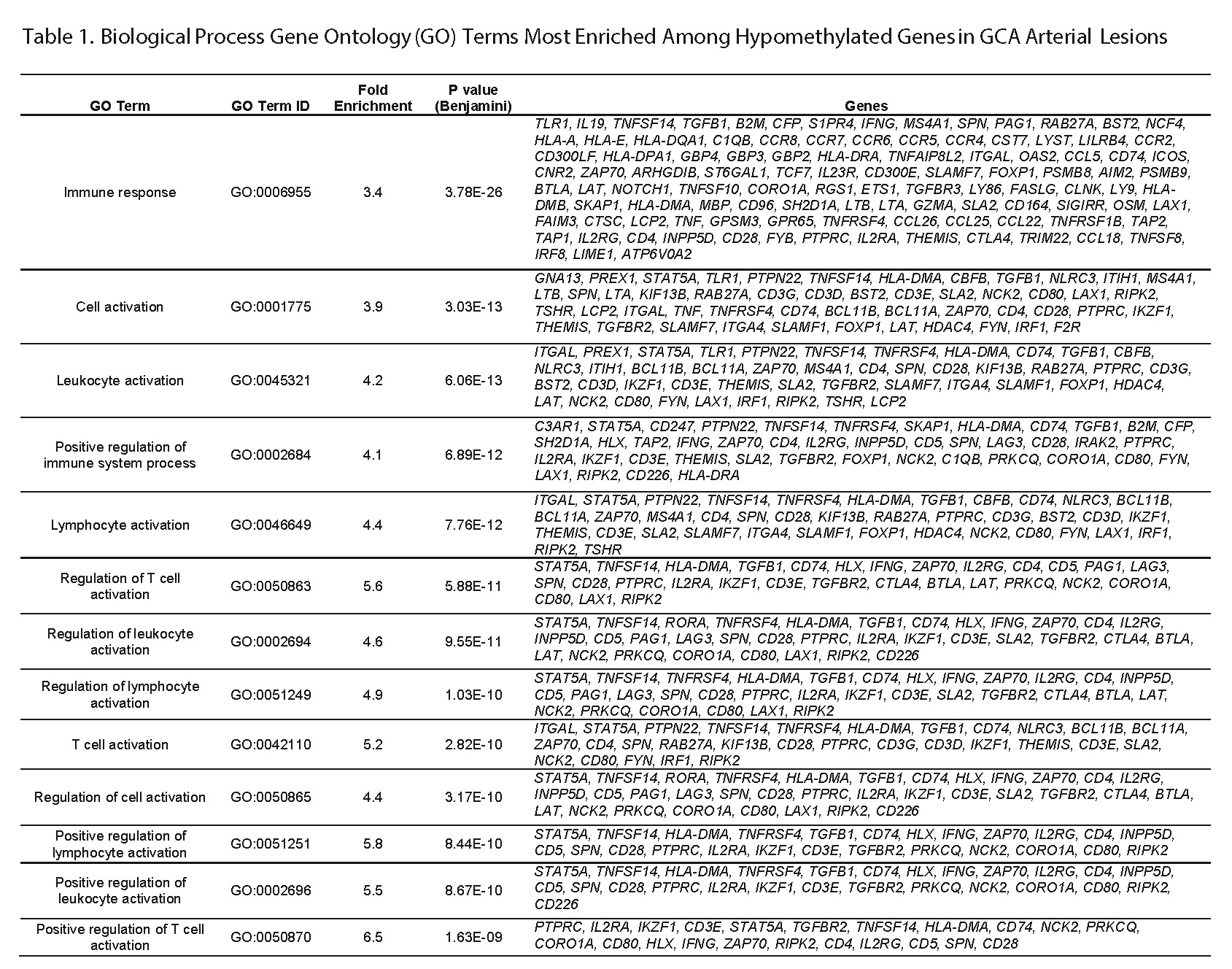
Results: We identified 1558 hypomethylated CpG sites (853 genes) and 2754 hypermethylated CpG sites (1471 genes) in GCA patients compared to controls. Gene ontology enrichment analysis of hypomethylated genes revealed significant representation in T cell activation and differentiation pathways, including both Th1 and Th17 related cytokine gene signatures (Table 1). Other proinflammatory genes such as TNF, LTA, and LTB were significantly hypomethylated in the cellular milieu of GCA arteries. Of the hypomethylated CpG sites in TNF, which is a known genetic susceptibility locus for GCA, two are within 1500bp upstream of the transcription start site, while two CpG sites are in the gene body. In addition, other genetic susceptibility loci for GCA such as IFNG, PTPN22, and NLRP1 were also hypomethylated in this study. CCR7 which is expressed on mature DCs, was amongst the most hypomethylated genes in GCA, consistent with a previously described pathogenic role for mature DCs within GCA affected arteries. Gene ontology analysis of hypermethylated genes displayed enrichment for genes related to actin cytoskeletal arrangement and regulation of GTPase-mediated and Ras protein signal transduction.
Conclusion: DNA methylation profiling in GCA affected arteries revealed a robust T cell signature and identified key molecules that might help to better understand the pathogenesis of GCA.
Disclosure:
P. S. Coit,
None;
L. B. De Lott,
None;
B. Nan,
None;
V. M. Elner,
None;
A. H. Sawalha,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/dna-methylation-analysis-of-the-temporal-artery-microenvironment-reveals-a-robust-t-cell-signature-and-suggests-a-role-for-tnf-%ce%b1-in-giant-cell-arteritis/