Session Information
Date: Saturday, November 16, 2024
Title: Abstracts: SLE – Diagnosis, Manifestations, & Outcomes I: Omics
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) and dermatomyositis (DM) are hypothesized to be triggered by shared genetic and environmental factors leading to aberrant activation of innate immune pathways such as type I interferons and neutrophil extracellular trap (NET) formation. Despite these similarities in pathogenesis, SLE and DM are distinctive clinical phenotypes with variable responses to the same therapeutic agents. The present study addresses the shared and distinct dysregulated pathways between SLE and DM by comparing peripheral blood transcript and protein expression in a combined multi-enrichment analysis of pathway and ligand-receptor signaling.
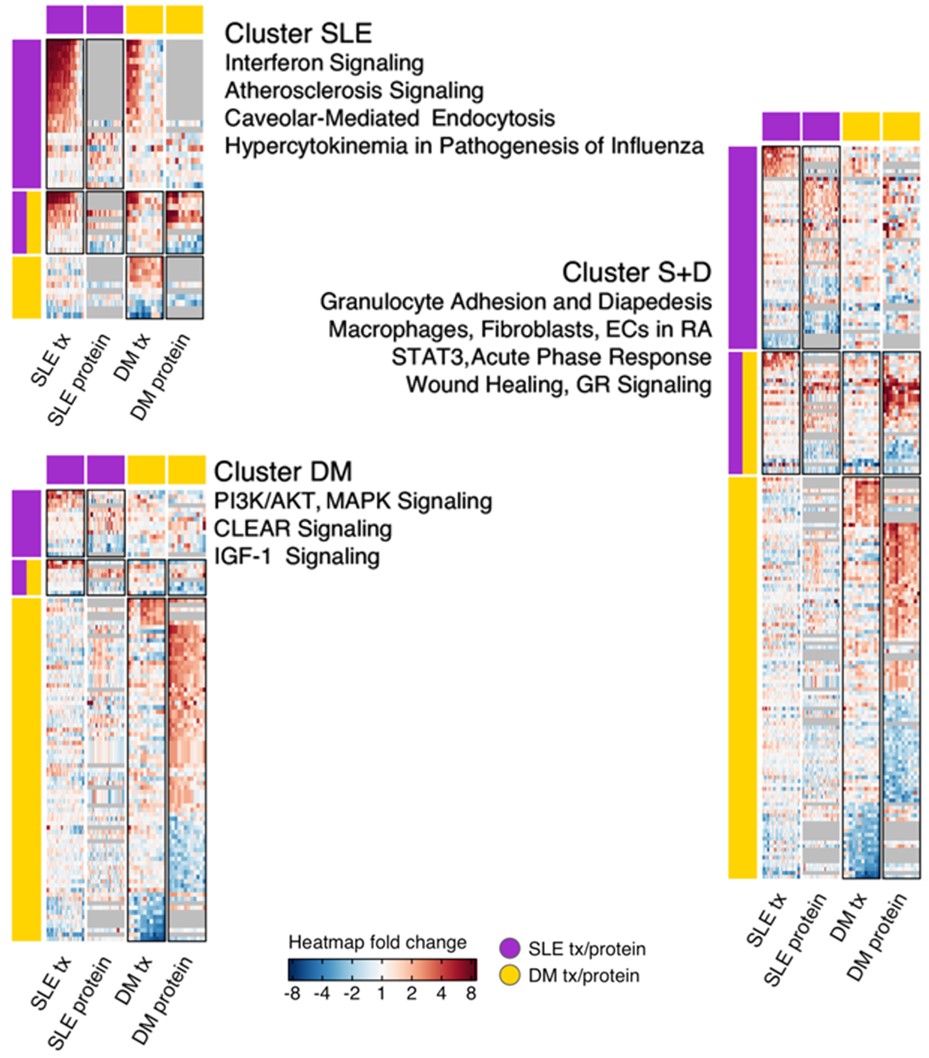
Methods: 48 SLE patients were age- and gender-matched 3:1 to 16 adult DM patients. 58 whole blood samples from sex- and age- (±10 years) matched healthy donors (HD) were collected. Whole blood RNA and sera were analyzed for transcriptomics and proteomics (using SomaLogic and RBM platforms). Significant transcript and protein results were combined across platforms into one collective set of gene locus abundance changes per patient cohort for pathway enrichment. Multi-enrichment analysis was performed using the top 10 pathways. Pathways were clustered using each gene-pathway incidence matrix, then split into four sub-clusters for within-cohort analyses and five sub-clusters for cross-cohort analyses.
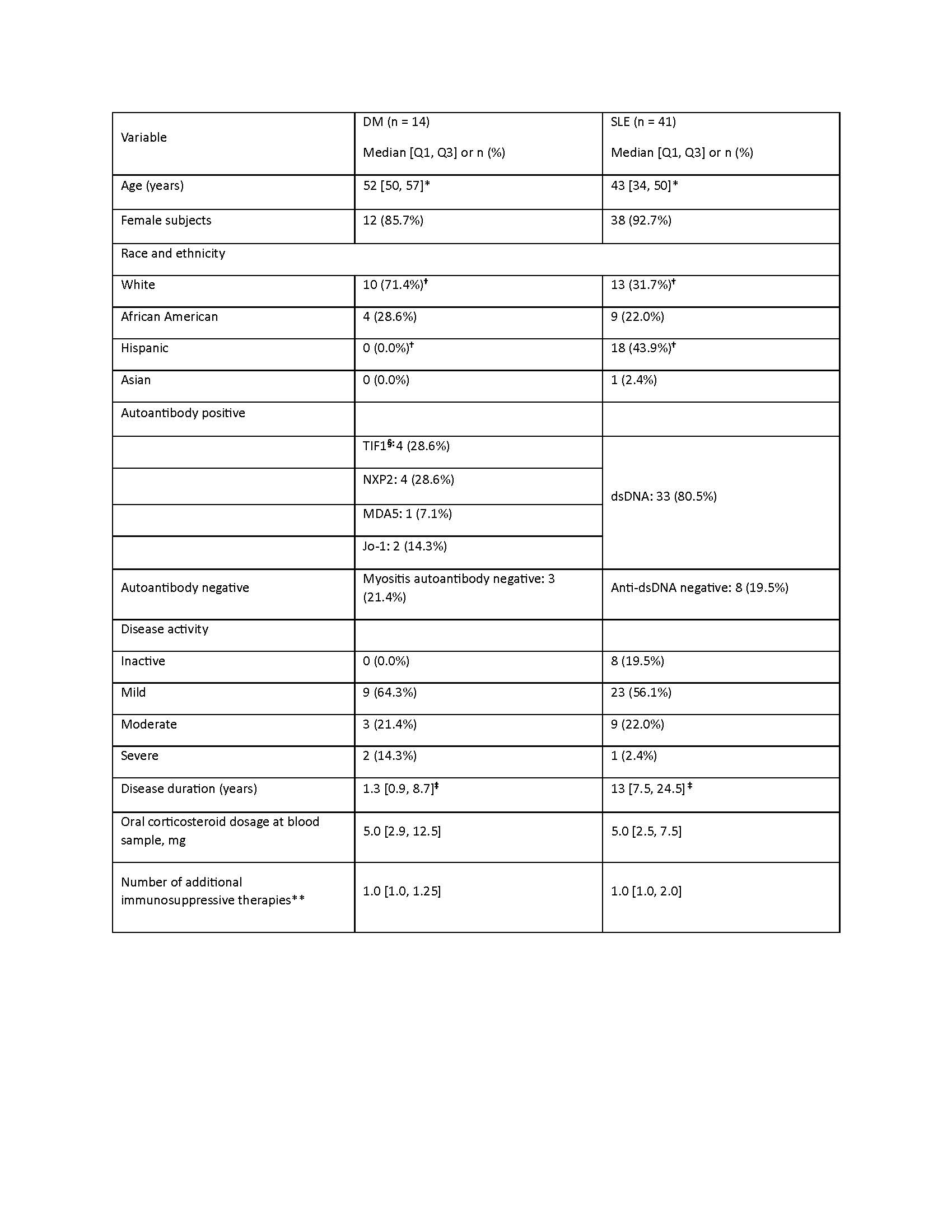
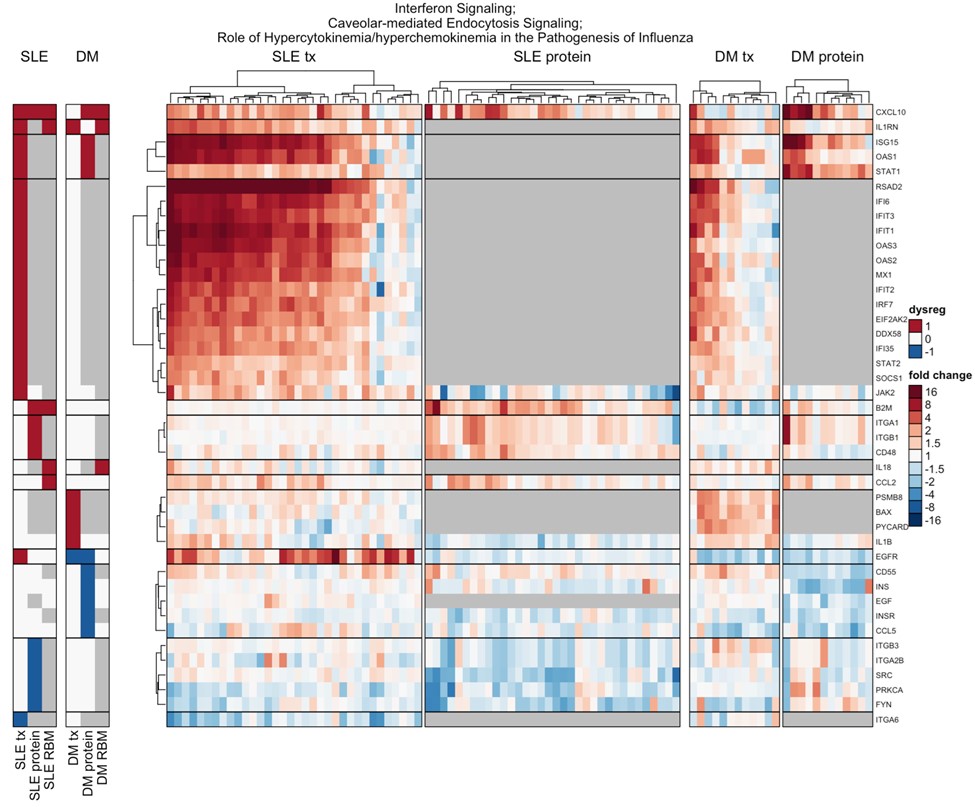
Results: After quality control, 41 SLE and 14 DM samples were analyzed for transcriptomics, and 34 SLE and 14 DM with matching HD were analyzed for proteomics. Baseline characteristics were similar, except DM patients were older, with shorter disease duration, and on higher doses of glucocorticoids (Table 1). SLE expression analysis produced 222 significantly dysregulated transcripts and 150 proteins. Twenty gene loci in SLE and 31 in DM were confirmed across platforms. Granulocyte Adhesion and Diapedesis, Glucocorticoid Receptor Signaling, Wound Healing Signaling, Hepatic Fibrosis, and Acute Phase Response Signaling pathways were significantly enriched in both SLE and DM, with 22% gene loci shared and 88% concordance in direction (Figure 1). SLE-specific pathways included Interferon, Hypercytokinemia in Pathogenesis of Influenza, and Atherosclerosis Signaling (Figure 2). DM-specific pathways included PI3K/AKT, IGF-1, coordinated lysosomal expression and regulation (CLEAR), and MSP-RON.
Conclusion: Our study revealed a moderate overlap in underlying pathogenic pathways between SLE and DM, with several pathways that were distinct for each. Both cohorts were also found to have unique, significantly enriched gene loci and proteins. These distinct dysregulated pathways may help explain differences in clinical phenotypes and responses to treatment.
The majority of SLE patients (27 of 34, 80%) show very high (8- to over 16-fold) up-regulation of interferon-activated genes, while only a subset of DM patients (4 of 12, 33%) show moderate (2- to 4-fold) up-regulation.
To cite this abstract in AMA style:
Ward J, Ambatipudi M, Manna Z, Smith M, de los Reyes M, Schiffenbauer A, Rahman S, Zerrouki K, Miller F, Kaplan M, Li J, Casey K, Rider L, Hasni S. Distinct Transcript and Protein Dysregulation Patterns in Dermatomyositis and Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/distinct-transcript-and-protein-dysregulation-patterns-in-dermatomyositis-and-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/distinct-transcript-and-protein-dysregulation-patterns-in-dermatomyositis-and-systemic-lupus-erythematosus/