Session Information
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Single cell transcriptional profiling (scRNA-Seq) is valuable in identifying gene signatures and cell subpopulations associated with rheumatoid arthritis (RA). However prior studies have often overlooked important demographic confounders and not focused on non-remission and difficult to treat RA. The aim of this study is to identify peripheral blood mononuclear cells (PBMCs) of RA and disease-activity cell subsets and gene signatures in a diverse population of patients.
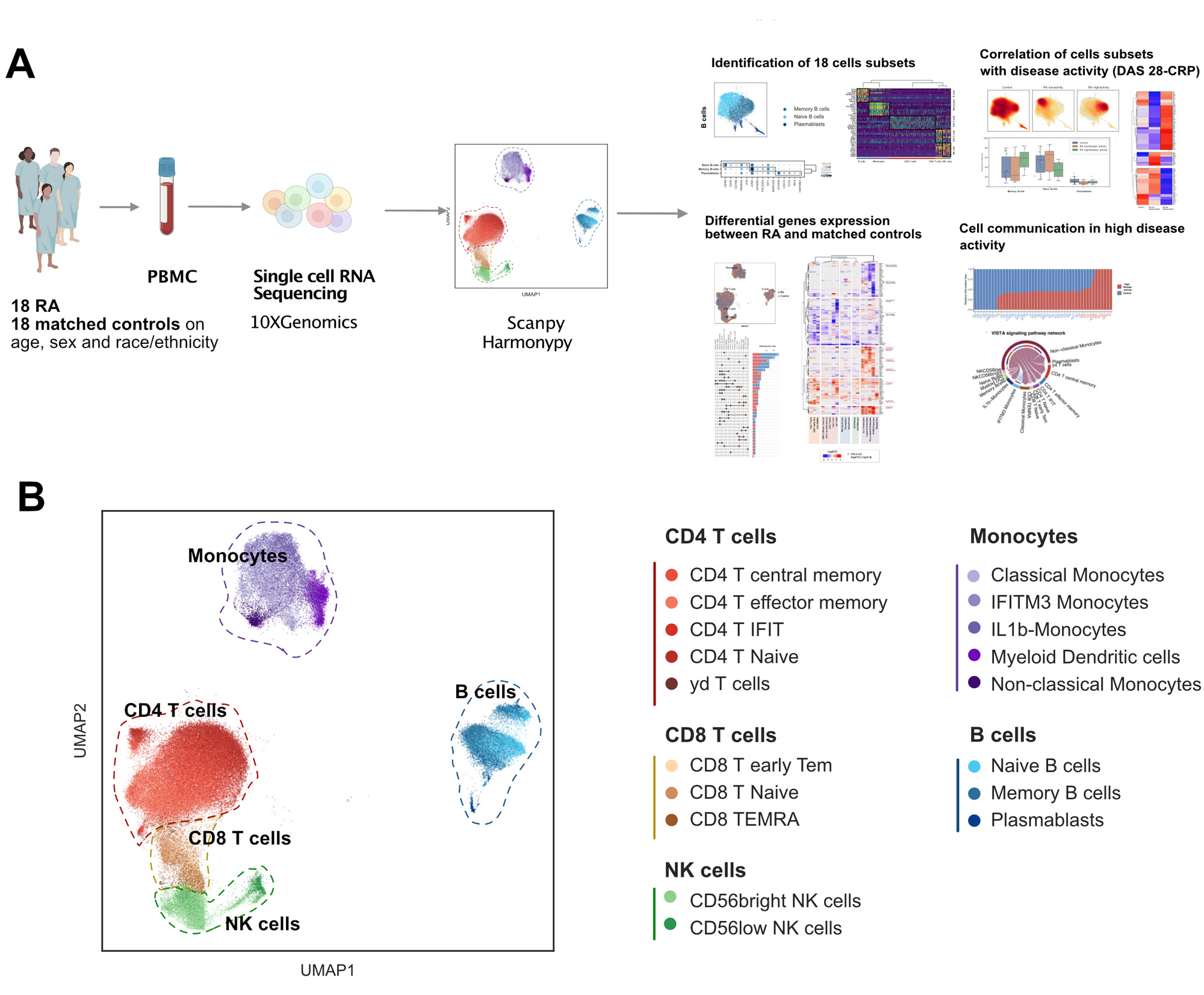
Methods: 10X Chromium single cell sequencing was performed on peripheral blood mononuclear cells (PBMC) from 18 early RA patients and 18 healthy controls matched on age, sex, race and ethnicity. Data were processed using standard CellRanger and Scanpy pipelines, with HarmonyPy for batch correction. Differential expression (DE) was computed using pseudobulk analysis and DESeq2. Pathway analysis was carried out with over-representation analysis (ORA). Mann-Whitney tests were used to assess differences in cell proportions between matched RA and control samples, as well as between RA patients with low disease activity or remission (DAS28-CRP< 3.2; n=9) versus moderate or high disease activity (DAS28 CRP≥3.2;n=7). Ligand-receptor interaction analysis was performed using Cellchatdb.
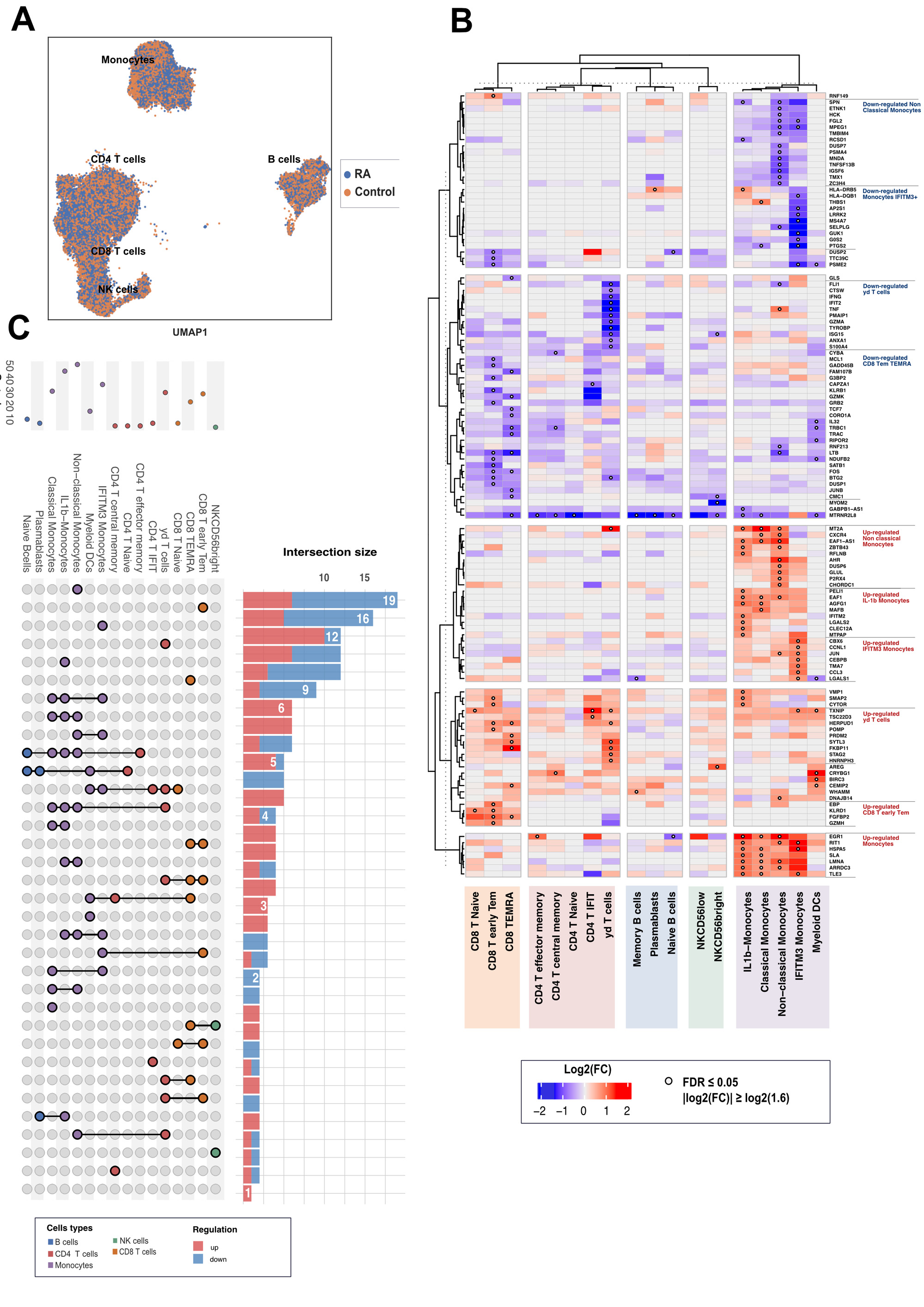
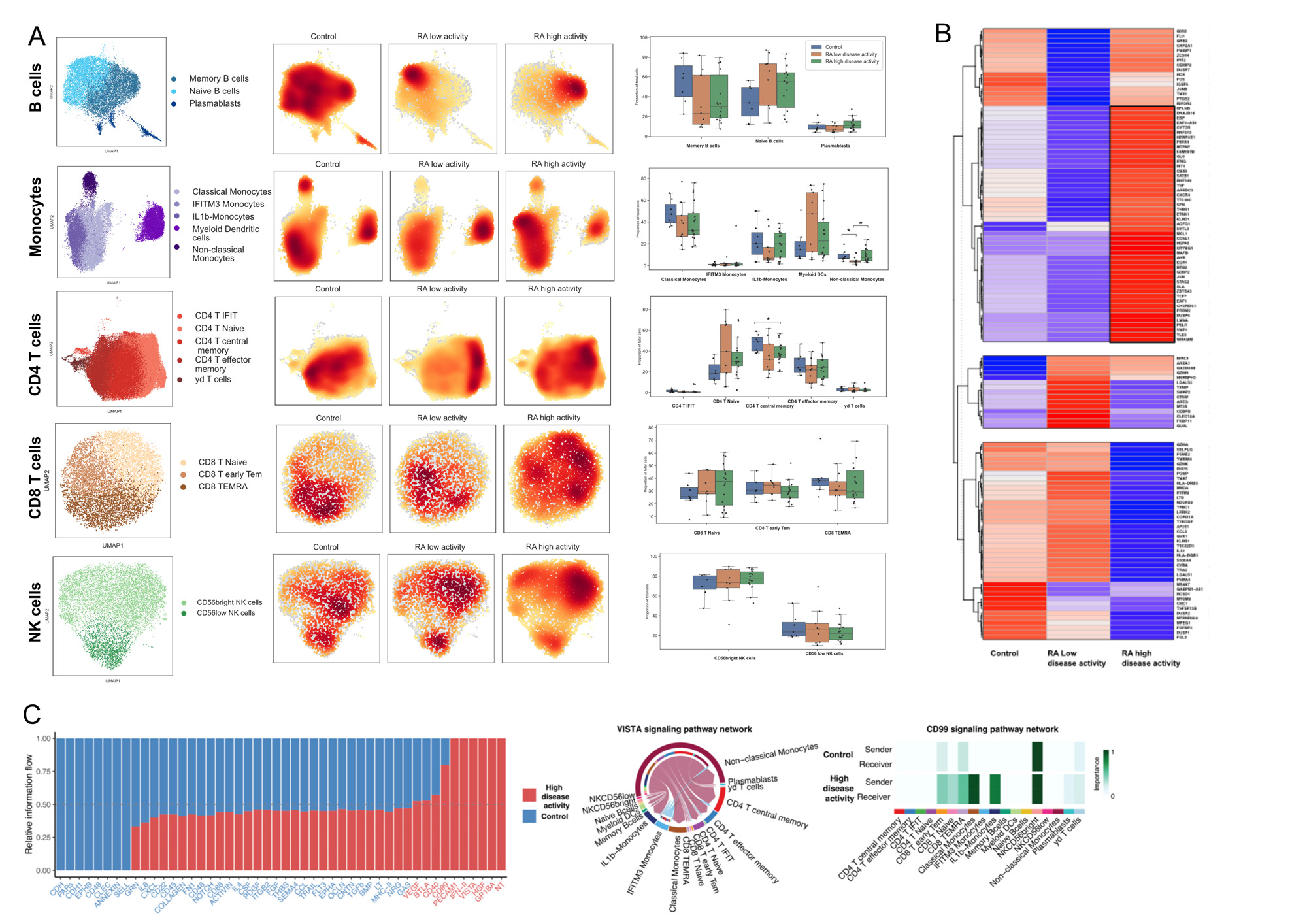
Results: The final dataset consisted of 22,159 genes across 125,698 cells. We identified 18 PBMC subsets, including 5 CD4+ T cell subsets, 3 CD8+ T cell subsets, 2 Natural Killer cell subsets, 3 B cell subsets, and 5 monocyte subsets (Figure 1). Within these subsets, IFIT CD4 T+ cells and IFITM3 monocytes were associated with Interferon-gamma response. 168 genes were DE between RA and matched controls (FDR≤0.05, foldchange >1.6). We identified up-regulation of pro-inflammatory genes associated with monocyte subsets and downregulation of inflammatory genes in gamma-delta T cells. Several genes associated with RA predisposition such as HLA-DRB5 and HLA-DQB1 were specifically downregulated in IFITM3 monocytes (Figure 2). Functional analysis and ORA highlighted significant enrichment of B cell activation and B cell receptor signaling pathways. Differences in cell subset proportions between patients with high and moderate activity, patients in low disease activity and remission, and healthy controls (n=18, Figure 3) were also observed. Non classical monocytes and T central memory were decreased in patients with high disease activity compared to control and high disease activity (p=0.022; p=0.034). We also identified a specific signature of 49 genes, including IFNG, TNF, KLRD, EGR1, CBX6, CXCR4, JUN, and TLE3, that was significantly associated with disease activity. Finally, cell communication analysis between patients with high disease activity and controls revealed upregulation of IFN-II, VEGF, VISTA, BTLA, and CD40 pathway signaling.
Conclusion: Here we describe a dataset of scRNA-Seq PBMCs from a diverse population of patients with RA and matched healthy controls. We identify differentially expressed genes and cell subsets linked to disease activity, providing insights into RA pathophysiology and potentially new therapeutic targets.
To cite this abstract in AMA style:
Binvignat M, Miao B, Wibrand C, Yang M, Rychkov D, Flynn E, Khan U, Nititham J, Carvidi A, Krueger M, Niemi E, Sun Y, Fragiadakis G, Klatzmann D, SELLAM J, Mariotti-Ferrandiz E, Gross A, Ye C, Butte A, Criswell L, Nakamura M, Sirota M. Deciphering Rheumatoid Arthritis Disease Activity-Associated Gene Signatures and Cell Subsets Through Single Cell Transcriptomics [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/deciphering-rheumatoid-arthritis-disease-activity-associated-gene-signatures-and-cell-subsets-through-single-cell-transcriptomics/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deciphering-rheumatoid-arthritis-disease-activity-associated-gene-signatures-and-cell-subsets-through-single-cell-transcriptomics/