Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Synovial fibrosis is a hallmark of osteoarthritis (OA) and a clinical correlate to joint pain. Fibrosis of skin, lungs and other organs is the hallmark of systemic sclerosis (SSc). Both OA synovium and SSc skin exhibit extracellular matrix changes, ectopic chondrogenesis/mineralization, nociceptive sensitization, and inflammatory infiltrates, but studies directly comparing these conditions are lacking. We utilized human single cell RNA sequencing (scRNAseq) data to interrogate the nature and extent of conserved fibroblast signaling in OA synovium and SSc skin, using idiopathic pulmonary fibrosis (IPF) as a comparison of bona fide fibrosis.
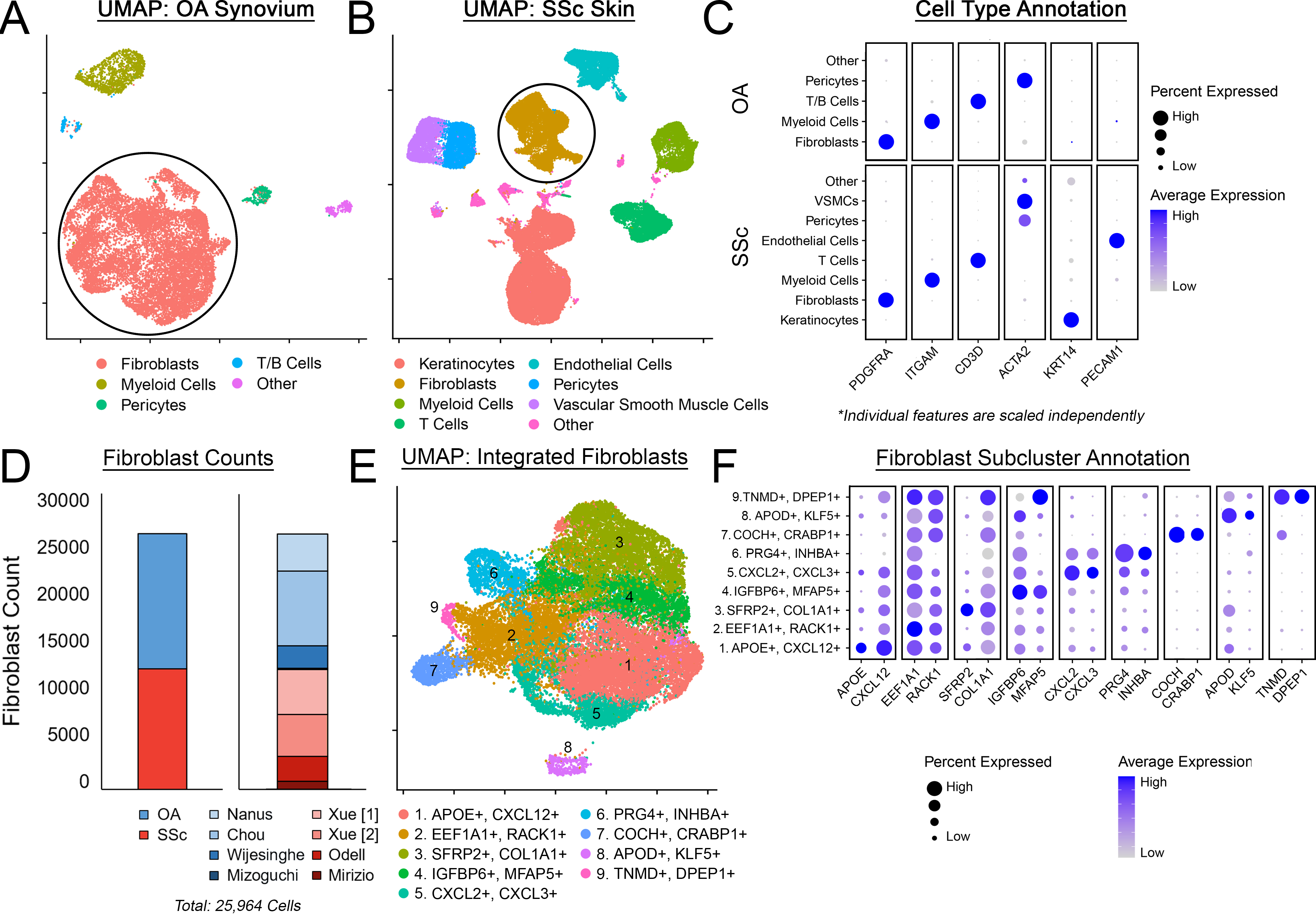
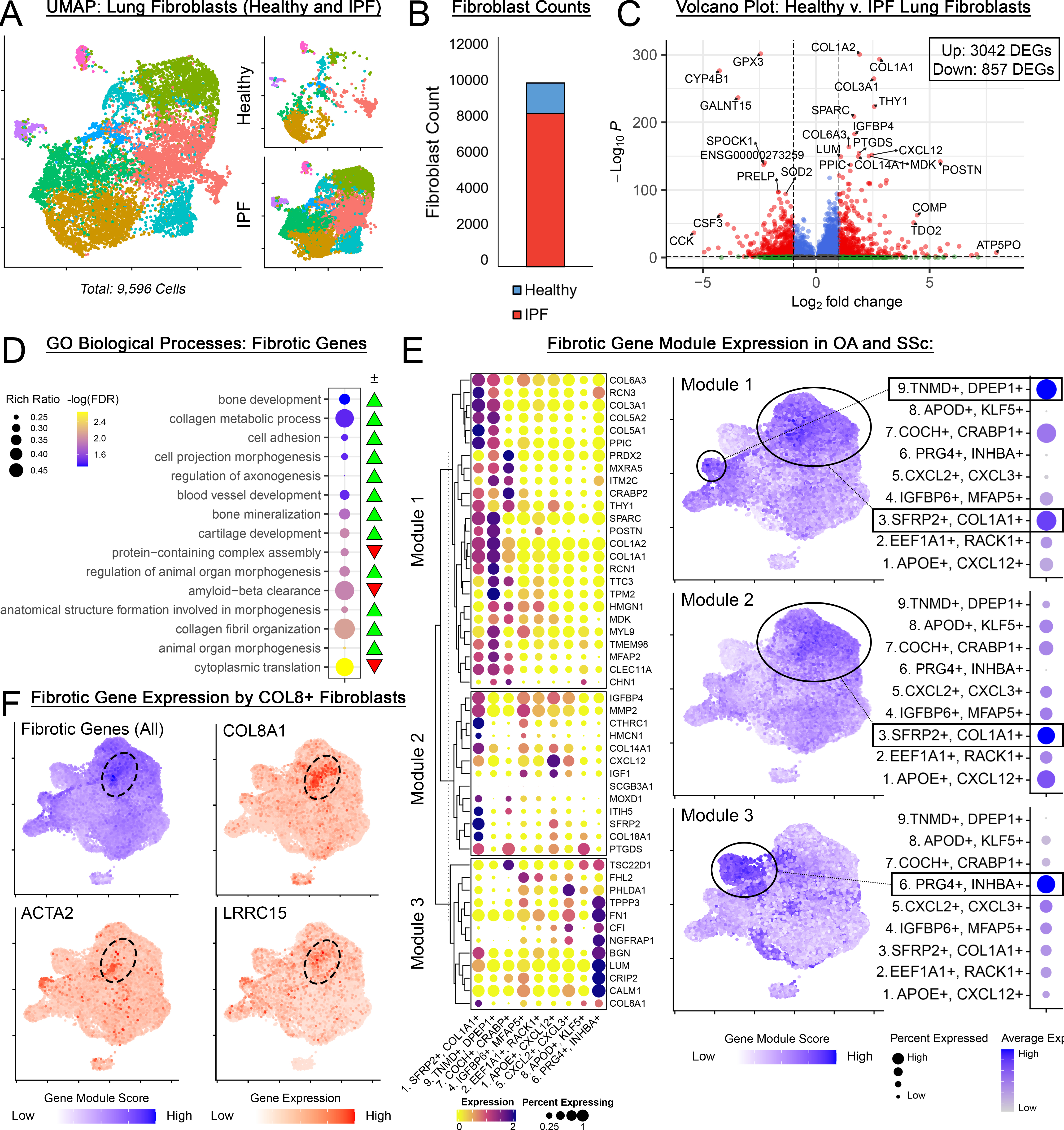
Methods: An integrative analysis utilized publicly available scRNAseq datasets from human OA synovium and SSc skin (Fig 1ABD). Major cell types were clustered and identified (Fig 1C). Fibroblasts from OA and SSc datasets were re-integrated into a joint data object and subclustered into 9 populations with distinct gene signatures (Fig 1EF). Subclusters were analyzed to determine relative abundance, cluster-specific gene markers, and functional roles (via gene ontology, GO). To define a “fibrotic reference gene set”, human lung scRNAseq data of healthy and IPF patients (N=6) was obtained (Fig 3AB) and differential gene expression was assessed between healthy and IPF fibroblasts (Fig 3C).
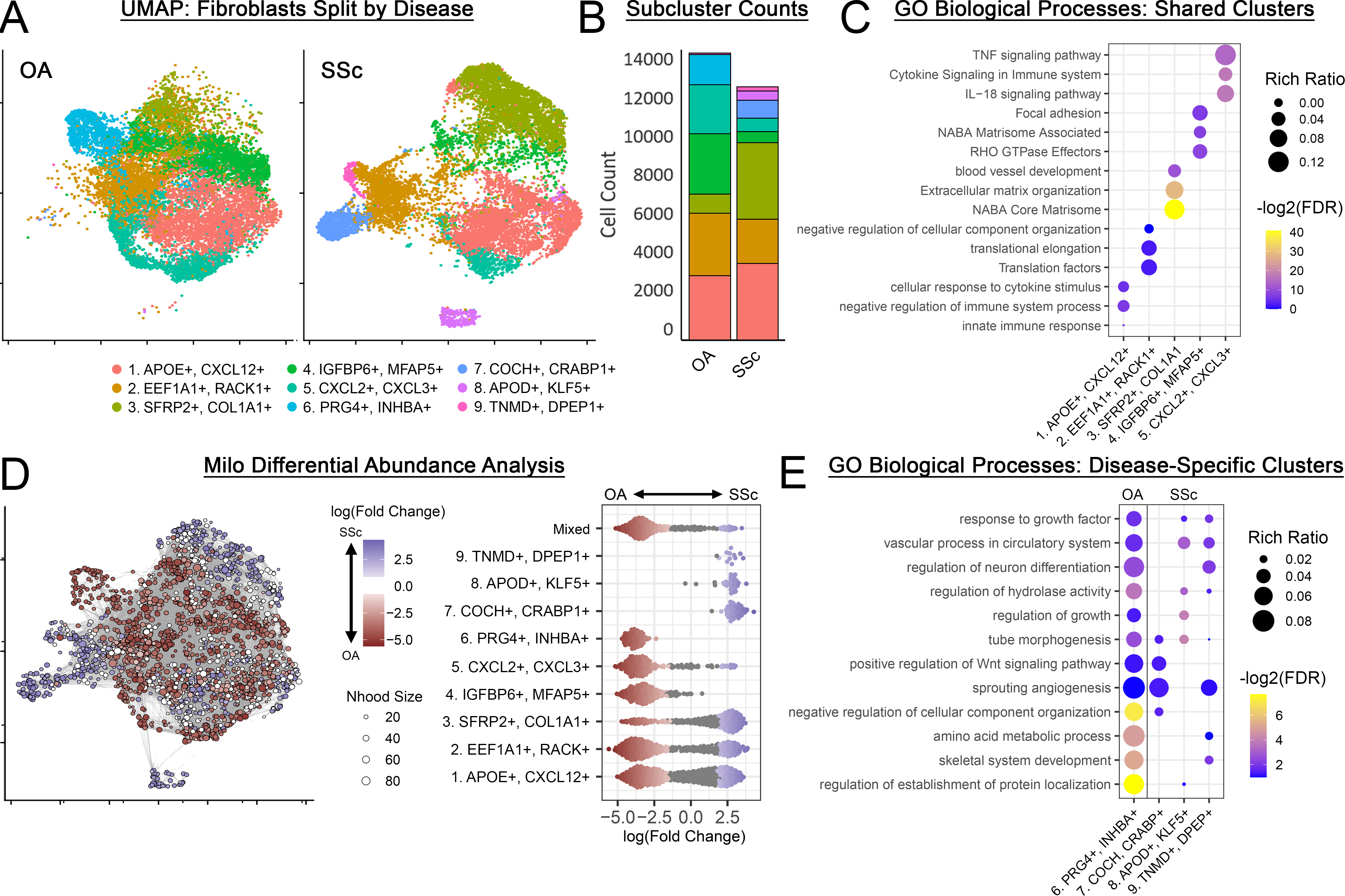
Results: Five fibroblast clusters (1,2,3,4,5) were substantially conserved in OA and SSc and participated in shared, disease-relevant GO terms (matrix deposition, immune/inflammatory signaling, Fig 2A-D). One cluster (6) was unique to OA (synovial lining fibroblasts); 3 clusters (7,8,9) were unique to SSc (Fig 2ABD). Despite being transcriptionally distinct, OA- and SSc- specific clusters both express genes related to shared, disease-relevant GO terms (Wnt signaling, neuromodulation, angiogenesis, Fig. 2E). A fibrotic reference gene set was identified (Fig 3C) comprising 3,899 genes differentially expressed between IPF and healthy lung fibroblasts, involved in fibrosis-relevant GO terms (collagen metabolism, chondrogenesis, mineralization, neuromodulation, Fig 3D). The top 50 genes overexpressed in IPF were clustered into 3 distinct modules – within OA/SSc fibroblasts, these modules were primarily expressed within 3 subclusters (Fig 3E), with strong module 1 expression in clusters 3 (SFRP2+) and 9 (TNMD+), module 2 in cluster 3 (SFRP2+), and module 3 in cluster 6 (PRG4+). Across all 481 genes strongly overexpressed in IPF (Padj < 0.05, log2fc > 1), the highest expression in OA/SSc fibroblasts occurred in a distinct subset of cells expressing COL8A1, which has been linked with cardiac fibrosis, along with myofibrotic markers (Fig 3F).
Conclusion: OA synovium and SSc skin share several transcriptionally-distinct fibroblast subsets related to matrix deposition and immunomodulation, suggesting these fibrosis-relevant functions are well-conserved between the two conditions. OA and SSc fibroblasts both express a fibrotic gene signature consistent with bona fide fibrotic disease (e.g. IPF). This expression is largely confined to specific fibroblast subsets, establishing potential therapeutic avenues targeting these subpopulations for fibrosis treatment.
To cite this abstract in AMA style:
Newton M, Mohan A, Hanna T, Orange D, Varga J, Maerz T. Conserved Pro-Fibrotic Fibroblast Signatures in Osteoarthritis Synovium and Systemic Sclerosis Skin [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/conserved-pro-fibrotic-fibroblast-signatures-in-osteoarthritis-synovium-and-systemic-sclerosis-skin/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/conserved-pro-fibrotic-fibroblast-signatures-in-osteoarthritis-synovium-and-systemic-sclerosis-skin/