Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Antiphospholipid syndrome (APS) is a systemic autoimmune disease characterized with the presence of pathogenic antiphospholipid antibodies (aPL) by autoreactive B cells. However, it remains vague that at which stages in the central-to-peripheral lineage of B cell in APS patients acquire autoreactivity.
Methods: To obtain in-depth understanding of immune tolerance disturbance in APS patients, we performed single-cell transcriptome (RNA) and immune repertoire (VDJ) sequencing on mononuclear cells of paired bone marrow (BMMC) and peripheral blood (PBMC) samples from 13 untreated primary APS patients, (5 thrombotic APS patients, 4 obstetric APS patients and 4 aPL-carriers) and 3 healthy donors.
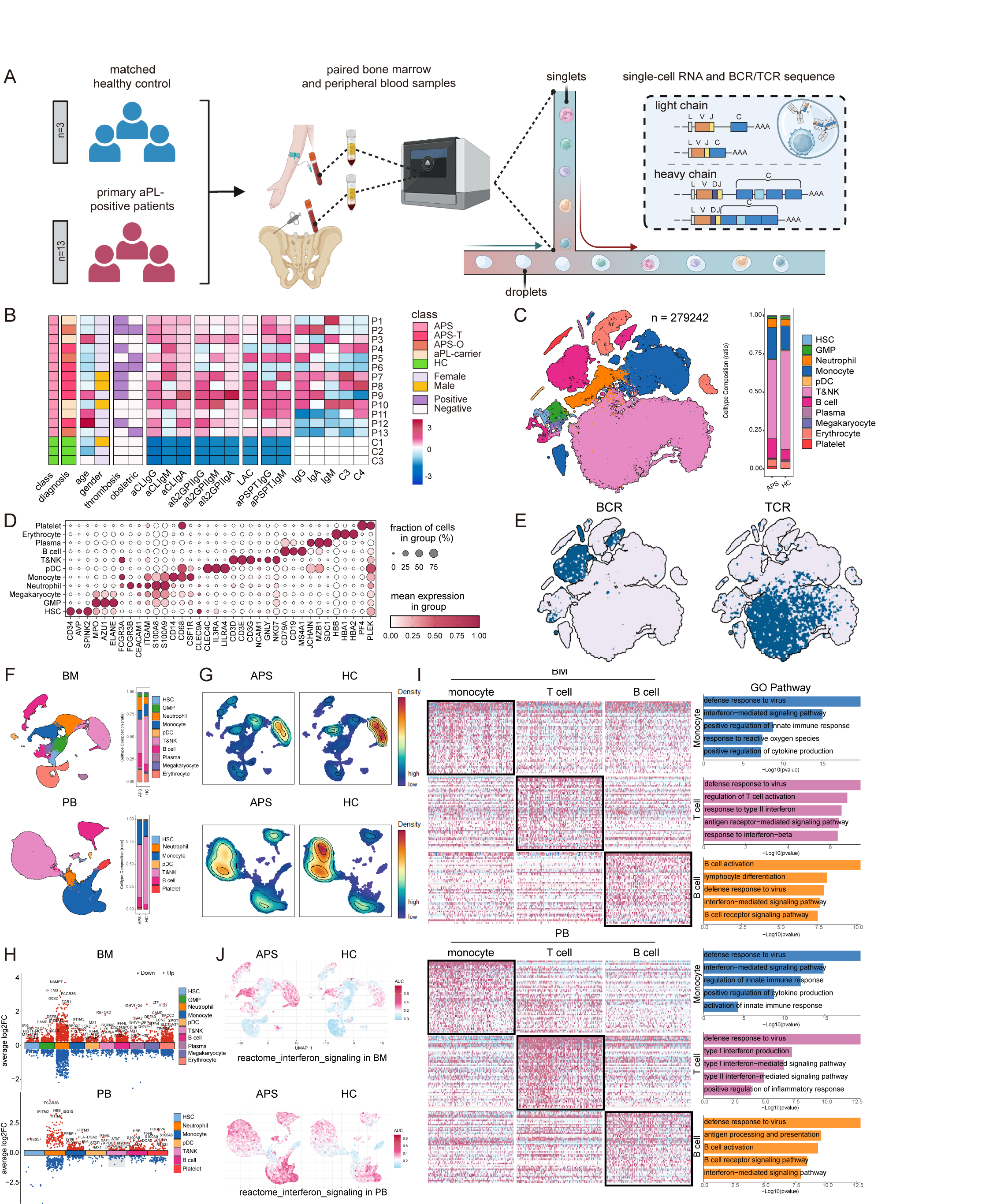
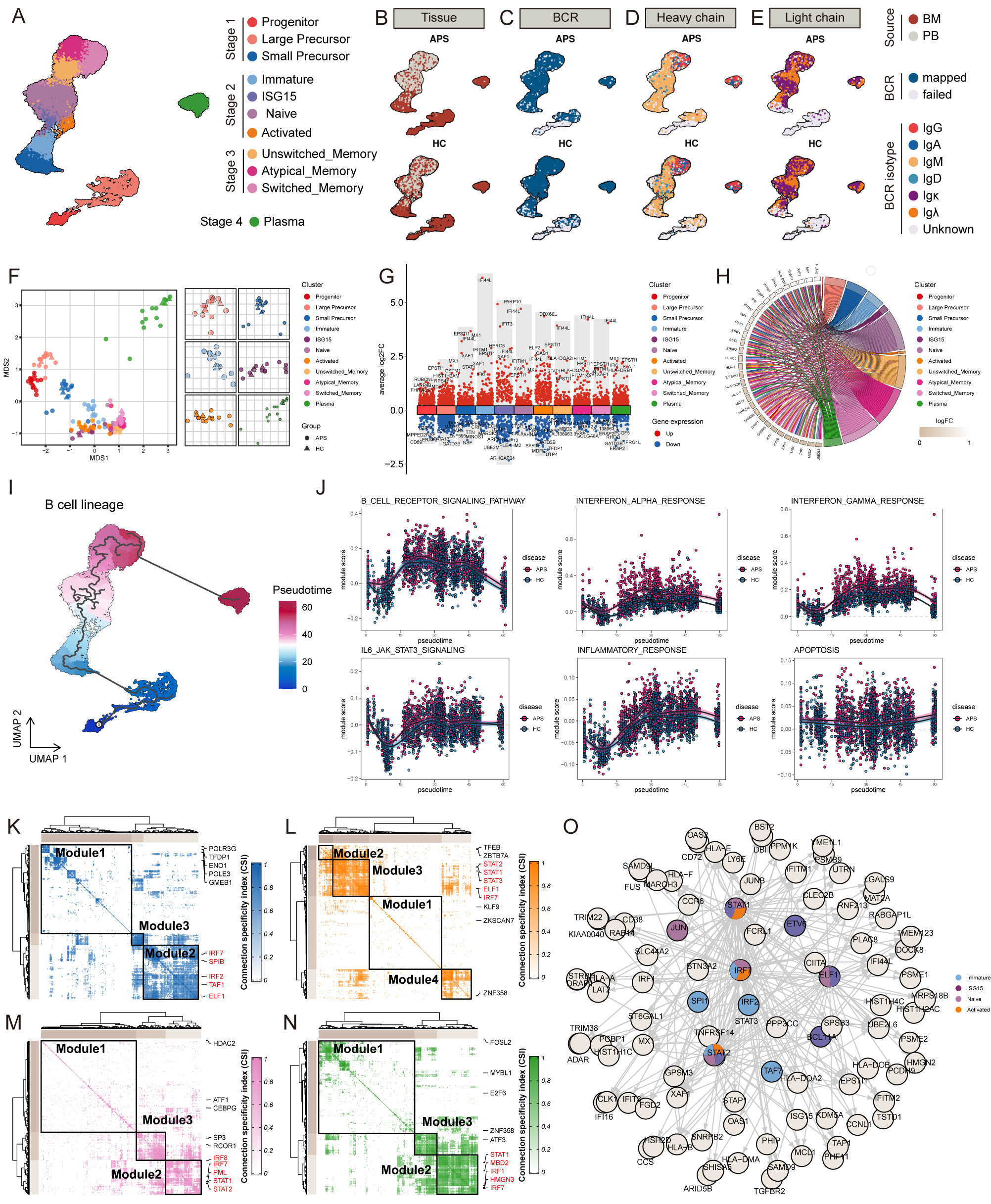
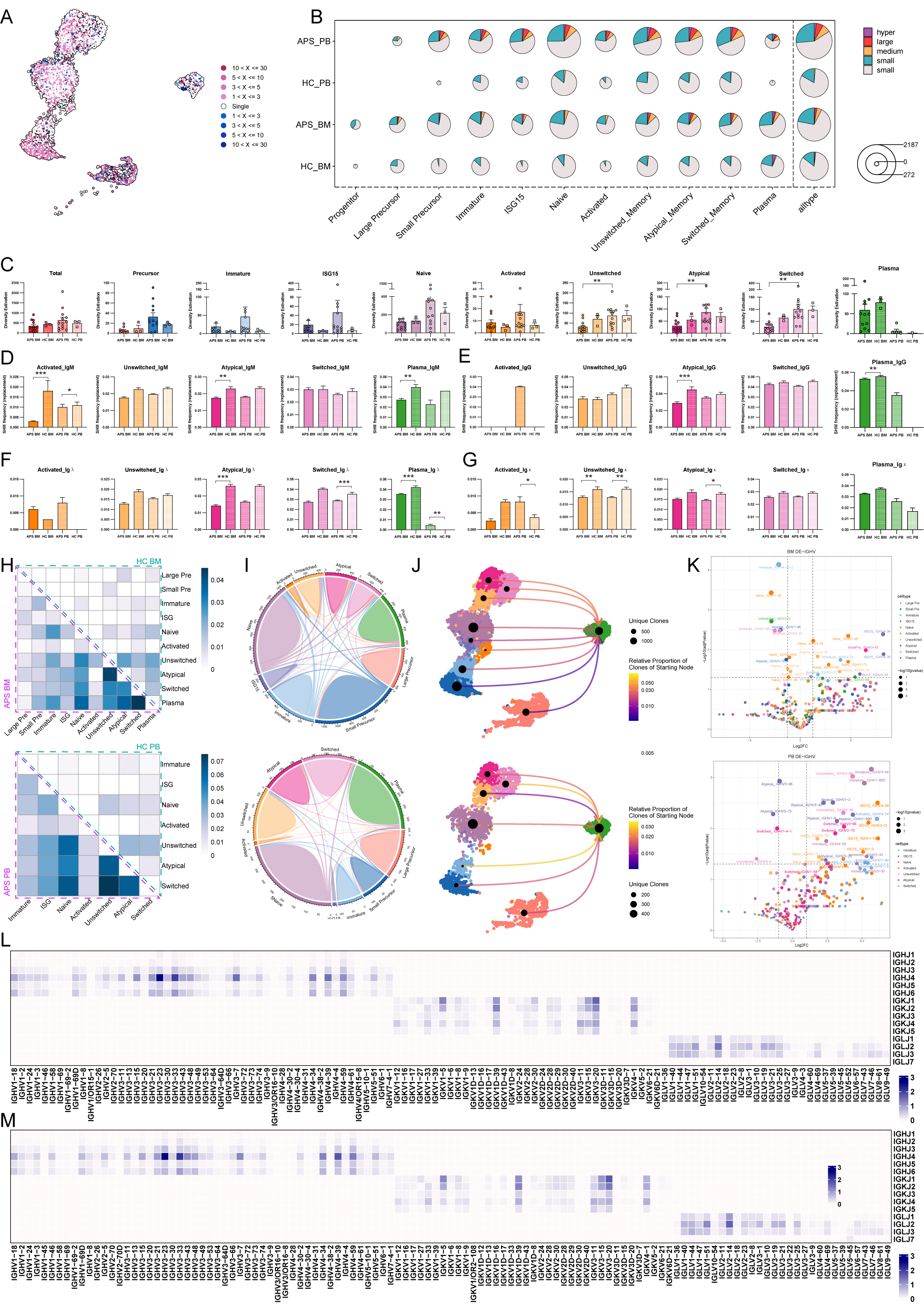
Results: After quality control and filtration, we obtained a total of 279,242 cells with integrated transcriptional and V(D)J features, including 113,843 BMMCs and 165,399 PBMCs. Analysis indicated widespread upregulation of interferon-associated genes (e.g., MX1, IFI6, IFITM3, IFI44L, IFI27) across multiple cell types in both BM and PB, with early-stage immune cells (HSC and GMP) also showing upregulated interferon-associated genes, suggesting foundational roles in shaping APS immune landscape (Figure 1). To further explore B cell abnormalities, we consolidated B cells and plasma cells from BM and PB to reconstruct the developmental lineage, identifying eleven distinct B cell subtypes categorized into four developmental stages. APS patients showed higher percentages of B cell subtypes and significant correlations between thrombotic events, B cell compositions, and anti-β2GP1 IgG titers, indicating quantitative abnormalities and progressive increases in B cell composition, especially in early-to-middle stages. Functional transcriptome analysis revealed pronounced differential states in early and intermediate B cells, with upregulation of interferon-related genes, antigen presentation genes, and key transcription factors. GO enrichment analysis highlighted significant aerobic respiration, cell cycle activities, and B cell signaling pathways. Elevated inflammatory and interferon responses were observed throughout B cell lineage in APS patients (Figure 2). Additionally, V(D)J sequencing data showed increased clonal expansion and clonal diversity in APS patients, particularly in early stages, with significant differences between BM and PB. Lower somatic hypermutation levels were noted in various B cell subsets, affecting high-affinity antibody production. BCR clonotype overlapping analysis demonstrated increased autoreactive potential, and differential usage of IGHV genes was enriched in APS patients. These findings describe abnormal BCR repertoire characteristics and underscore disturbed immune surveillance and central role of interferon-mediated pathways in APS autoimmunity (Figure 3).
Conclusion: We presented a comprehensive immune-desregulation blueprint in primary APS patients, as well as a significant influence of interferon pathway and profound understanding of the dynamics of obtaining autoreactivity alongside B cell lineage. These findings provide valuable insights into the emerging process of autoreactivity and potential therapeutic targets for APS.
To cite this abstract in AMA style:
Pan H, Wei X, Qian J, Yu S, Yang Z, Ding Z, Yang C, Shi H. Comprehensive Single-cell Analysis Reveals Interferon Pathway Activation and Aberrant B Cell Dynamics in APS Autoimmunity [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/comprehensive-single-cell-analysis-reveals-interferon-pathway-activation-and-aberrant-b-cell-dynamics-in-aps-autoimmunity/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/comprehensive-single-cell-analysis-reveals-interferon-pathway-activation-and-aberrant-b-cell-dynamics-in-aps-autoimmunity/