Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Juvenile dermatomyositis (JDM) and childhood-onset systemic lupus erythematosus (cSLE) are pediatric autoimmune diseases that can present with overlapping clinical features yet have unique tropism for major organ involvement. The objective of this study was to characterize differences in immune cell proportions and transcriptional signatures in treatment naïve (TN) JDM vs. cSLE to identify disease-specific mechanisms and treatment targets.
Methods: We isolated fresh peripheral blood mononuclear cells (PBMCs) from TN JDM (n=5), TN cSLE (n=4) and healthy controls (CTL; n=4). All patients met 2017 EULAR/ACR classification criteria for JDM or 2019 ACR/EULAR classification criteria for SLE, respectively. Single cell RNA sequencing was performed on the 10x Genomics platform. Ambient RNA and doublets were removed by SoupX and scDblFinder, respectively. We performed batch correction using Harmony, dimensionality reduction using principal component analysis and uniform manifold approximation and projection, and clustering using shared nearest neighbor. Cell types were annotated via published cell type specific marker genes. We analyzed (1) relative abundance of each cell type within disease and control by testing pair-wise significance using z-test and (2) differential transcriptomic signatures per cell type between JDM and cSLE by using likelihood ratio test and reported log fold change (logFC) and Bonferroni corrected p-values.
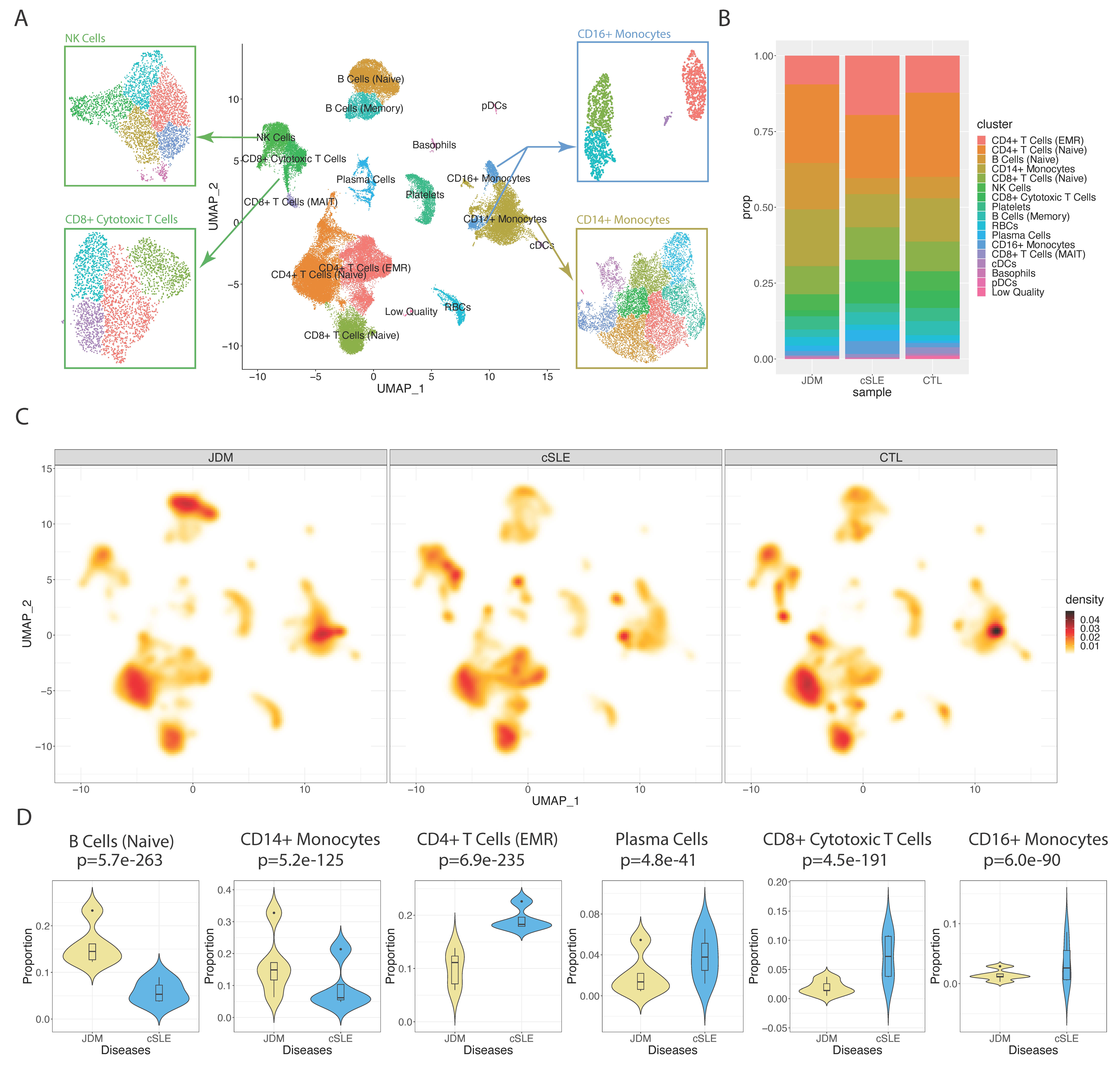
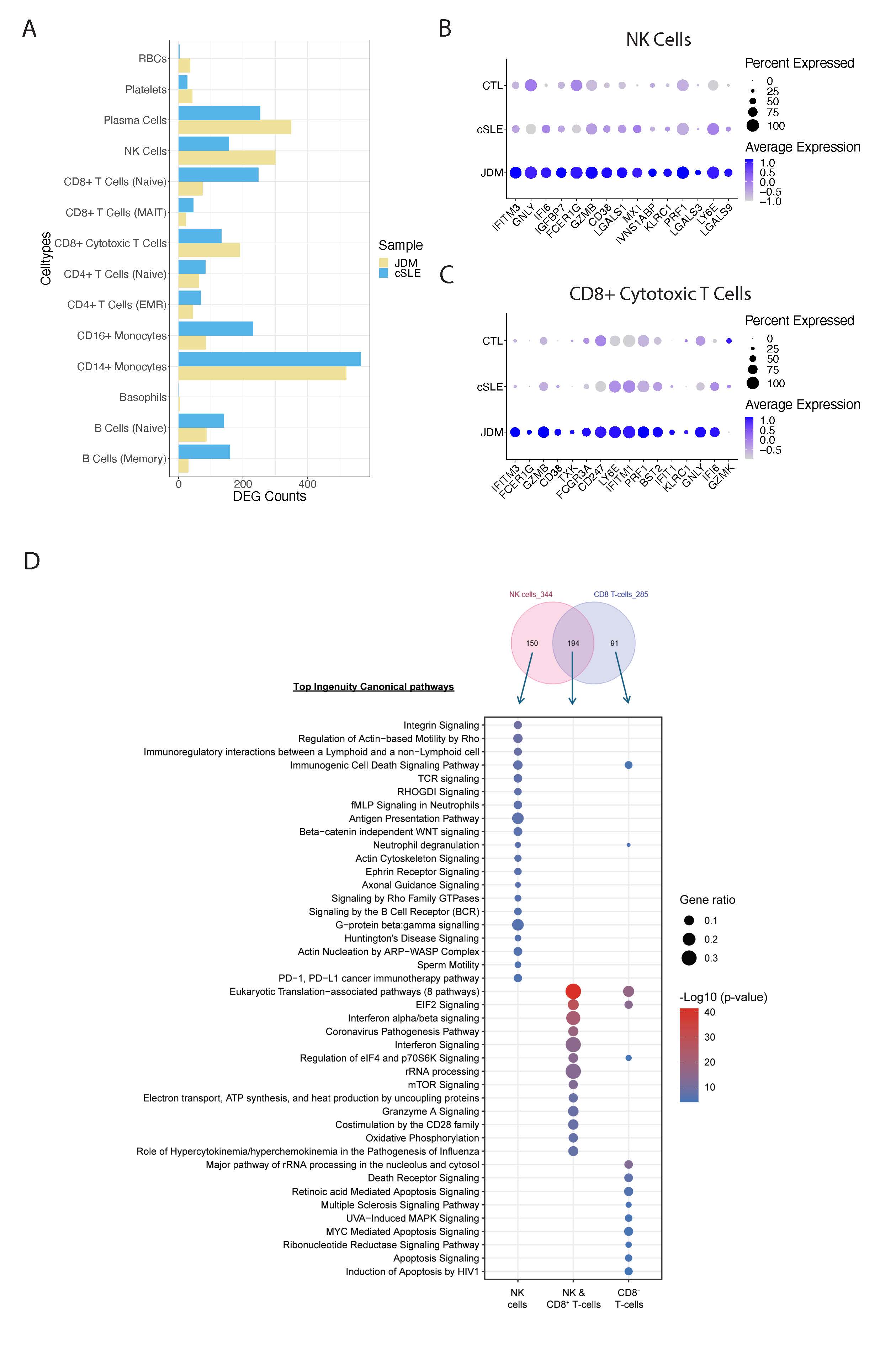
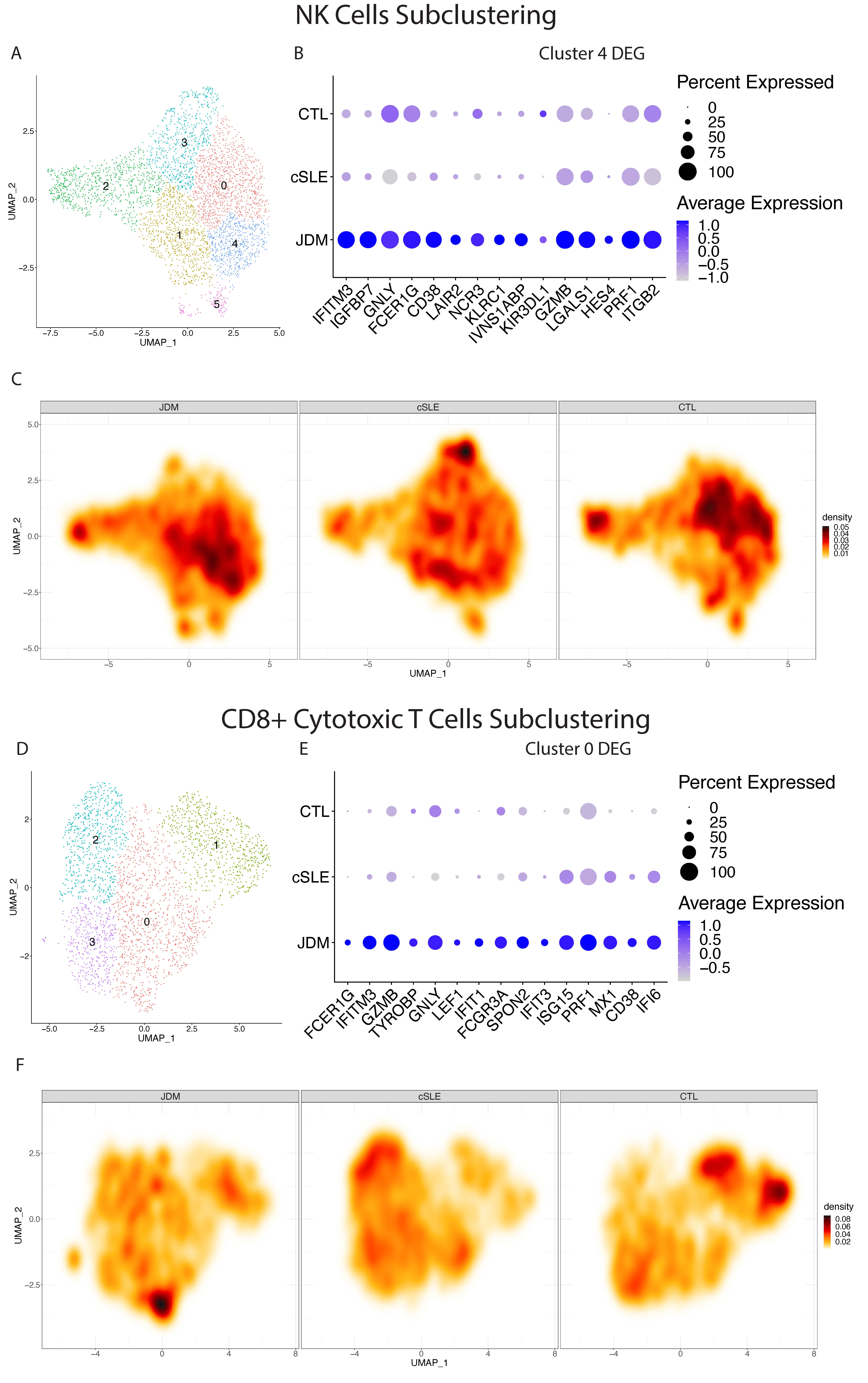
Results: 16 distinct annotated cell types were identified from 67,661 cells (Fig 1A). As compared to cSLE, JDM demonstrated increased relative abundance of naïve B-cells and CD14+ monocytes and decreased relative abundance of CD4+ effector memory and regulatory (EMR) T-cells, plasma cells, CD8+ cytotoxic T-cells and CD16+ monocytes (Fig 1B-D). Cytotoxic cells in JDM, including NK cells and CD8+ cytotoxic T cells, displayed a higher number of DE genes (DEG) as compared to cSLE (Fig 2A). JDM cytotoxic cells displayed a common increased cytotoxic and IFN signature, with 194 shared DEG, including GNLY, IFITM3, FCER1G, GZMB and CD38 in the top 10 DEG (Fig 2B-D). Interestingly, JDM CD8+ cytotoxic T-cells had increased GZMB expression as compared to increased GZMK in cSLE (Fig 2C). NK cells in JDM were uniquely enriched for antigen presentation, PD-L1 signaling and fMLP signaling in neutrophils, and CD8+ cytotoxic cells demonstrated enrichment for death receptor and apoptosis signaling (Fig 2D). Subclustering of cytotoxic cells highlighted an increase in NK subcluster 4 in JDM (Fig 3A & 3C), which had higher expression of immunoregulatory receptors of cytotoxicity, including NCR3, KLRC1 and KIR3DL1 (Fig 3B). JDM also had more cells in CD8+ cytotoxic T-cell subcluster 0 (Fig 3D & 3F), which was notable for increased expression of TYROBP, an adaptor protein mediating activation of multiple immune cell types (Fig 3E).
Conclusion: We identified differences in immune cell proportions in TN JDM vs. cSLE. We highlight cytotoxic cells in JDM vs. cSLE as displaying higher IFN, apoptotic and chemotactic signaling. Further investigation between JDM and cSLE-specific single-cell transcriptomic signatures has the potential to identify more specific treatments and biomarkers for each disease.
To cite this abstract in AMA style:
Li Q, Berthier C, Goudsmit C, Matossian S, Wasikowski R, Gudjonsson J, Kahlenberg J, Tsoi L, Turnier J. Circulating NK and CD8+ Cytotoxic T Cells in Treatment Naïve JDM Demonstrate Higher Cytotoxic and Interferon Signature as Compared to Childhood-Onset SLE [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/circulating-nk-and-cd8-cytotoxic-t-cells-in-treatment-naive-jdm-demonstrate-higher-cytotoxic-and-interferon-signature-as-compared-to-childhood-onset-sle/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/circulating-nk-and-cd8-cytotoxic-t-cells-in-treatment-naive-jdm-demonstrate-higher-cytotoxic-and-interferon-signature-as-compared-to-childhood-onset-sle/