Session Information
Session Type: Poster Session D
Session Time: 8:30AM-10:30AM
Background/Purpose: Early detection of psoriatic arthritis (PsA) in psoriasis (Pso) patients is crucial for timely treatment and prevention of structural joint damage. We aimed to identify disease-specific immune profiles discriminating Pso from PsA patients, potentially facilitating adequate referral.
Methods: The phenotype of PBMCs of consecutive Pso (N=45) and PsA (N=41) patients was determined using multi-color flowcytometry and univariate analysis to compare percentages of cell subsets between Pso and PsA. Disease-specific immune profiles were defined by machine learning using a random forest (RF) classification model. 1) The dataset was randomly split into a training (70%) and test-set (30%). 2) An RF model consisting of 1000 forests was built and optimized with internal cross-validation. 3) Step 1 and 2 were repeated 500 times. This procedure resulted in 500 RF-based models, each of which was evaluated by an Area under the Curve (AUC) analysis (Figure 1A/1B).
Results: In-depth immune phenotyping resulted in over one hundred different cell subsets for each blood sample being evaluated. A number of cell subsets was significantly different in Pso vs PsA. Key PsA identifying immune cell subsets selected by the RF classification model (AUC value of 0.95) included increased proportions of differentiated CD4+CD196+CD183–CD194+ and CD4+CD196–CD183–CD194+ T-cells and reduced proportions of CD196+ and CD197+ monocytes, memory CD4+ and CD8+ T-cells and CD4+ regulatory T-cells (Figure 1C shows top 10 PsA/Pso-classifying immune cell populations). Within PsA, joint scores showed a strong association with memory CD8+CD45RA-CCR7- effector T-cells and CD197+ monocytes (Figure 2A/2B).
Conclusion: Through the integration of an extensive phenotyping of blood immune cell subsets and a machine learning approach, we identified an immune profile which discriminates PsA from Pso. This immune profile may facilitate timely diagnosis of PsA in Pso and it highlights the possibility of using a combination of selected immune cell subsets as a method for detecting PsA in Pso patients.
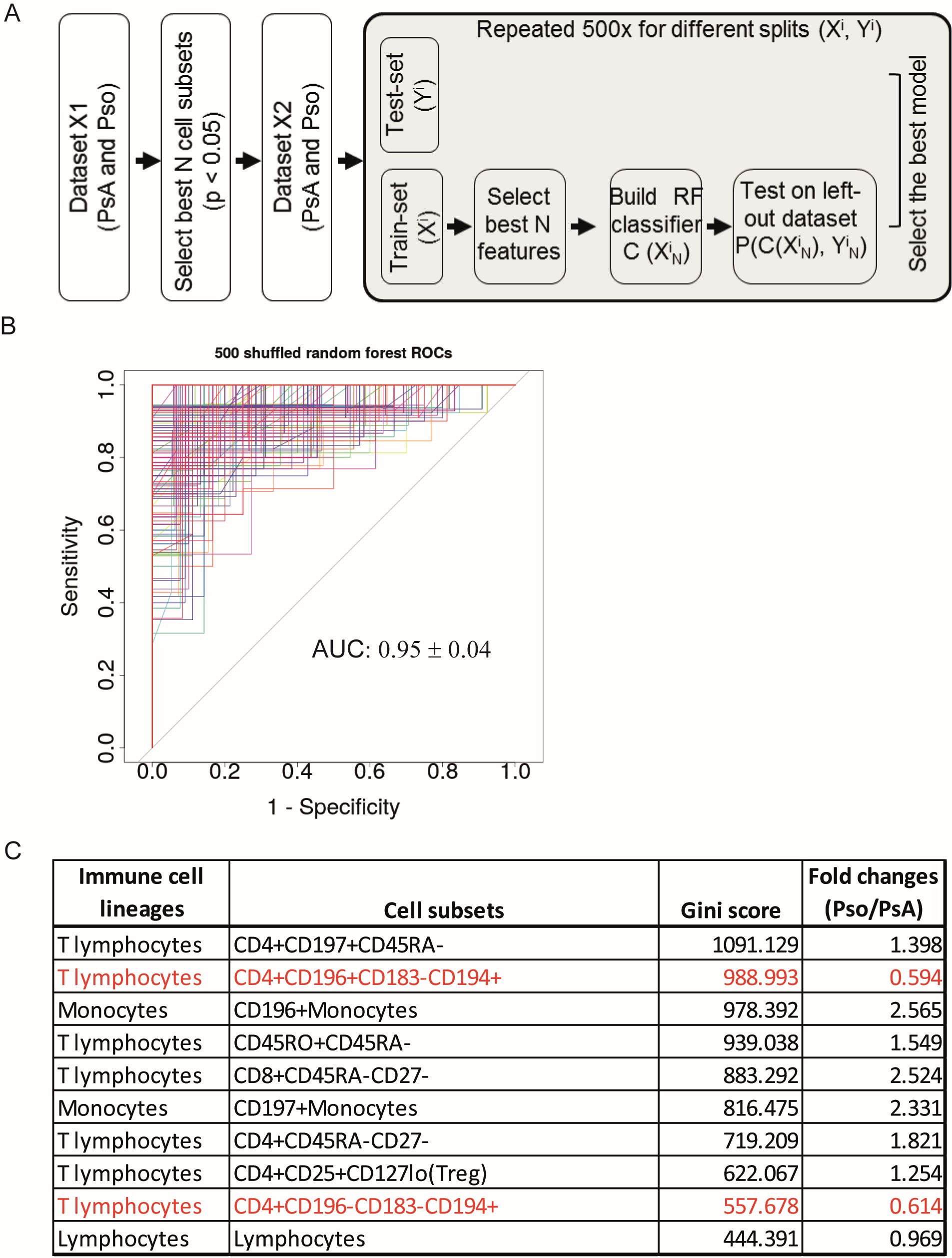 Figure 1: Discrimination of PsA from Pso patients using a random forest classification model. (A) Schematic overview of the data analysis procedure. The significantly different cell subsets in PsA vs Pso were selected based on the univariate analysis as shown in Fig 1A and the highly correlated cell subsets were excluded. Randomly splitting for training X’ (70%) and test Y’ (30%) dataset was repeated 500 times. The RF classification model (containing 1000 forests) was built using the training dataset and the test dataset was shuffled to cross-validate the model. (B) Overview of the ROC curves derived from 500 RF classification models. The AUC value was shown as mean ± SD. (C) Top 10 most relevant-cell subsets contributing to the classifications of PsA and Pso. Cell subsets were ranked based on gini-score. Fold change is computed as the ratio of mean values of each cell subset’s percentage in Pso vs PsA. Names of ICS in red indicate these cells are higher in PsA than in Pso.
Figure 1: Discrimination of PsA from Pso patients using a random forest classification model. (A) Schematic overview of the data analysis procedure. The significantly different cell subsets in PsA vs Pso were selected based on the univariate analysis as shown in Fig 1A and the highly correlated cell subsets were excluded. Randomly splitting for training X’ (70%) and test Y’ (30%) dataset was repeated 500 times. The RF classification model (containing 1000 forests) was built using the training dataset and the test dataset was shuffled to cross-validate the model. (B) Overview of the ROC curves derived from 500 RF classification models. The AUC value was shown as mean ± SD. (C) Top 10 most relevant-cell subsets contributing to the classifications of PsA and Pso. Cell subsets were ranked based on gini-score. Fold change is computed as the ratio of mean values of each cell subset’s percentage in Pso vs PsA. Names of ICS in red indicate these cells are higher in PsA than in Pso.
 Figure 2: Correlation of clinical parameters with PsA-classifying immune cells. (A) Heatmap showing the pearson’s correlation between percentages of cell subsets with clinical parameters in PsA. Numbers indicate the Pearson’s correlation coefficient, * p < 0.05; ** p < 0.01; *** p < 0.001. (B) Scatter plots showing the pearson’s correlation of the percentages of CD197+ monocytes and CD8+CD45RA-CD27- memory T-cells with swollen joint count 28/66 and Psoriatic Arthritis Disease Activity Score (PASDAS).
Figure 2: Correlation of clinical parameters with PsA-classifying immune cells. (A) Heatmap showing the pearson’s correlation between percentages of cell subsets with clinical parameters in PsA. Numbers indicate the Pearson’s correlation coefficient, * p < 0.05; ** p < 0.01; *** p < 0.001. (B) Scatter plots showing the pearson’s correlation of the percentages of CD197+ monocytes and CD8+CD45RA-CD27- memory T-cells with swollen joint count 28/66 and Psoriatic Arthritis Disease Activity Score (PASDAS).
To cite this abstract in AMA style:
Mulder M, He X, van den Reek J, Urbano P, Kaffa C, Wang X, van Cranenbroek B, van Rijssen E, van den Hoogen F, Joosten I, Alkema W, de Jong E, Smeets R, Wenink M, Koenen H. Blood-based Immune Profiling Combined with Machine Learning Discriminates Psoriatic Arthritis from Psoriasis Patients [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/blood-based-immune-profiling-combined-with-machine-learning-discriminates-psoriatic-arthritis-from-psoriasis-patients/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/blood-based-immune-profiling-combined-with-machine-learning-discriminates-psoriatic-arthritis-from-psoriasis-patients/
