Session Information
Date: Saturday, May 20, 2017
Title: Plenary Abstract Session 3
Session Type: Abstract Submissions
Session Time: 2:30PM-3:00PM
Background/Purpose: Juvenile Idiopathic Arthritis (JIA) comprises a heterogeneous group of conditions that share chronic arthritis as a common characteristic. International uniformity in classifying JIA, based predominantly on clinical characteristics at onset, has helped 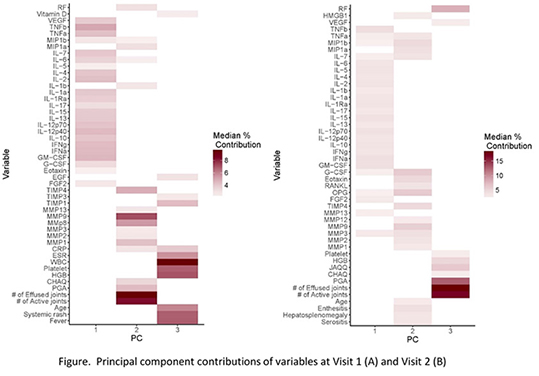 propel collaborative efforts to improve understanding of subset-specific pathophysiology, treatment responses, and outcomes. The purpose of this study was to consider the added value of combining biomarker-based attributes with clinical characteristics to classify chronic childhood arthritis in a biologically-based context.
propel collaborative efforts to improve understanding of subset-specific pathophysiology, treatment responses, and outcomes. The purpose of this study was to consider the added value of combining biomarker-based attributes with clinical characteristics to classify chronic childhood arthritis in a biologically-based context.
Methods: Data were derived from a prospective, nation-wide, longitudinal cohort study titled Biologically-Based Outcome Predictors in JIA (The BBOP Study). Newly diagnosed, treatment naïve children with JIA were evaluated at baseline and after six months. Data included clinical manifestations and plasma inflammation-related biomarkers. Categorical (nonlinear) principal component analysis (CAT-PCA), factor analysis of mixed data (FAMD), and probabilistic principal component analysis (PPCA) were used for dimensionality reduction purposes. To identify groups in the data, K-medoids clustering and Gaussian mixture modeling (GMM) were applied. Four common metrics were used to validate the clustering configurations: Davies–Bouldin, Dunn2, average silhouette width, and Calinski and Harabasz indices. The results were compared with the JIA subgroups defined by International League of Associations for Rheumatology criteria.
Results: A total of 150 JIA patients were enrolled. Data consisted of 191 variables. PCA reduced variables into 3 clinically relevant principal components (PCs) (Figure). Using PCs, three clusters were identified at baseline by both methods. At six months, three clusters were identified by K-medoids, and GMM recognized five clusters; new clusters were also revealed at six months (Figure). PCs recovered 33% and 47% of variance in the patient profiles in visit 1 and 2, respectively. Clustering validation indices showed that PPCA-GMM is the most reliable clustering method. At first presentation, clusters revealed in this analysis exposed different and more homogenous subgroups compared to the seven JIA ILAR subgroups. A large subset of patients with oligoarthritis and rheumatoid factor negative polyarthritis grouped into one cluster.
Conclusion: Using data-driven, unsupervised machine learning algorithms these analyses recognized distinctive patterns that provide insight into the underlying biology of chronic childhood arthritis and enable categorization of disease based on a combination of clinical and biomarker profiling.
To cite this abstract in AMA style:
Rezaei E, Hogan D, Trost B, Kusalik A, Benseler S, Boire G, Cabral DA, Cameron B, Campillo S, Chédeville G, Dancey P, Duffy CM, Watanabe Duffy KN, Ellsworth J, Eng S, Feldman BM, Gordon J, Guzman J, Houghton K, Huber A, Morris Q, Lang B, Levy DM, Matheson L, Oen K, Petty R, Ramsey S, Roth J, Rumsey D, Saint-Cyr C, Schneider R, Scuccimarri R, Silverman E, Spiegel LR, Stringer E, Tse SML, Tucker L, Yeung RSM, Rosenberg A. Biologically-Based Approach for Classifying Chronic Childhood Arthritis [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 4). https://acrabstracts.org/abstract/biologically-based-approach-for-classifying-chronic-childhood-arthritis/. Accessed .« Back to 2017 Pediatric Rheumatology Symposium
ACR Meeting Abstracts - https://acrabstracts.org/abstract/biologically-based-approach-for-classifying-chronic-childhood-arthritis/
