Session Information
Session Type: Poster Session B
Session Time: 8:30AM-10:30AM
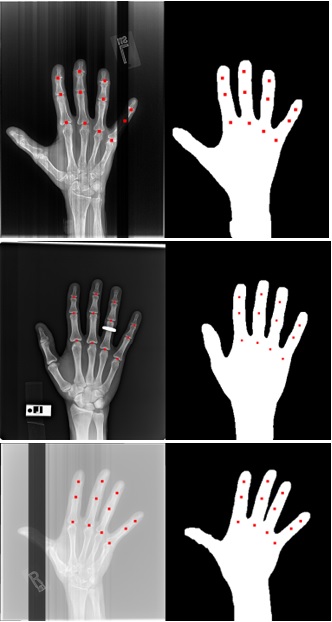
Background/Purpose: In many hand osteoarthritis (OA) studies (e.g., automatic Joint Space Width measuring), accurate hand segmentation is the first step towards making further analysis. However, it would be a difficult task to use only traditional image processing techniques due to inconsistencies like bands, rings, and low contrast X-rays (see Figure 1). To train a machine learning model to segment hand X-rays, one needs to set up a training set in which each hand X-ray should be manually delineated. To minimize the effort of manual delineation, we developed a novel iterative training strategy that requires only a small number of images to be manually segmented.
Methods: 3557 hand X-rays from the Osteoarthritis Initiative (OAI) were utilized in this study, where each X-ray contains one hand. We randomly separated the dataset into a training set with 3024 X-rays and a testing set with 533 X-rays, and the testing set was not exposed to the models during the training procedure. In round 1, we applied a traditional image segmentation algorithm on the training set and selected 132 good-quality results as a training set to train model 1. Round 2, we applied to model 1 to the training set with 3024 images and selected 25 of the worst predicted segmentations from the results. We manually segmented those 25 failed images and used them to train a new U-net model (model 2). In round 3, likewise, we applied to model 2 to the training set again and picked 11 failed segmentations from the results. We manually segmented those 11 failed cases and combined them with the 25 cases from round 2 to train a new model using a total of 36 training samples (model 3). In total, we only manually segmented 36 hand X-rays (25 in round 2 and 11 in round 3). To evaluate segmentation accuracy, two research assistants manually marked the centers of 12 joints (excluding the thumb, Figure 1 red dots) on each of the 533 hands in the testing set. The reason that we used joint centers instead of segmenting the whole hand is to save labor. We evaluated the hand segmentation accuracy at the joint level: if a joint center is within the predicted hand mask, the joint is a match; otherwise, the joint is a miss. The overall accuracy of a model is equal to the number of matches divided by the number of total joints.
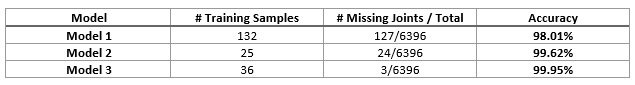
Results: Table 1 lists the accuracy for each model: model 1 performed the initial segmentation with the most training samples, but its accuracy was lowest; model 2 used only 25 training samples but improved the accuracy significantly; model 3 achieved the best accuracy (99.95%) with 36 training samples and only 3 joints out of 6396 joints were missed in the testing set.
Conclusion: Acquiring a large medical image dataset is a challenge in many studies and manually labeling is often a time-consuming procedure. In this paper, we proposed an iterative strategy to minimize the training samples that need to be manually labeled. In a dataset of 3557 hand X-rays, we only need to manually segment 36 images and achieved 99.95% accuracy for joint detection on the testing set. This strategy has potential usage in other medical image processing problems to save the manual labeling effort (e.g., knee cartilage segmentation, bone marrow lesion segmentation, etc.).
To cite this abstract in AMA style:
Yang Z, Shan J, Guida C, Blackadar J, Cheung T, Driban J, McAlindon T, Zhang M. Automatic Hand Segmentation from Hand X-rays Using Minimized Training Samples and Machine Learning Models [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/automatic-hand-segmentation-from-hand-x-rays-using-minimized-training-samples-and-machine-learning-models/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/automatic-hand-segmentation-from-hand-x-rays-using-minimized-training-samples-and-machine-learning-models/