Session Information
Date: Wednesday, November 13, 2019
Title: 6W023: Systemic Sclerosis & Related Disorder – Clinical III: Predictors of Outcome (2912–2917)
Session Type: ACR Abstract Session
Session Time: 11:00AM-12:30PM
Background/Purpose: Systemic sclerosis (SSc), or scleroderma, is a heterogeneous disease that is divided into limited cutaneous (lcSSc) and diffuse cutaneous (dcSSc) forms based on the extent of skin involvement, and internal organ involvement also adds to phenotypic variability. The three main internal organ complications are interstitial lung disease (ILD), pulmonary arterial hypertension (PAH) and scleroderma renal crisis (SRC).
Methods: We investigated classical HLA alleles and polymorphic amino acids for association with clinical subsets of SSc. A conditional association analysis with a Bonferroni-corrected significance threshold of P-value (P) < 0.00005 identified multiple subset-specific SSc risk factors in an African American cohort of 934 SSc patients and 946 unaffected controls.
Results: HLA-DRB1*08:04, an African-ancestry predominant allele, was the strongest risk factor with odds ratios (ORs) ranging from 2.7 to 3.3-fold in the lcSSc, dcSSc, ILD+ and PAH+ subsets of SSc. The allele HLA-DPB1*13:01 also independently increased risk in the dcSSc and ILD+ subsets, with ORs of 2.0 (95% CI 1.4-2.8) and 2.5 (95% CI 1.8-3.6), respectively. There were no significant associations in the SRC+ subset at the chosen P threshold, but the strongest association was with HLA-DQB1*03:19 (P = 0.0002), which is in linkage disequilibrium with HLA-DRB1*08:04. For the first time, a protective effect of HLA-DRB4*01:01 was identified in SSc, with a stronger effect in the lcSSc subset (OR = 0.4, 95% CI 0.4-0.7) than in the dcSSc subset (OR = 0.5, 95% CI 0.4-0.7). The presence of this allele is also linked to HLA-DRB1*07:01, which has been reported as protective for SSc in European ancestry cohorts. Further stratification of clinical subsets by autoantibody status revealed more striking effects of certain alleles, despite the reduced sample size. On comparing the dcSSc with the lcSSc none of the alleles met the statistical significance threshold due to reduction in sample size. Interestingly, the top two associations were both class I HLA alleles, HLA-A*74:01 (OR = 0.5, 95% CI 0.3-0.8) and HLA-C*17:01 (OR = 1.7, 95% CI 1.1-2.6), and these associations became even stronger upon comparing patients with dcSSc to those with CREST syndrome (OR = 0.4 and 2.0 and P = 0.0008 and 0.003, respectively).
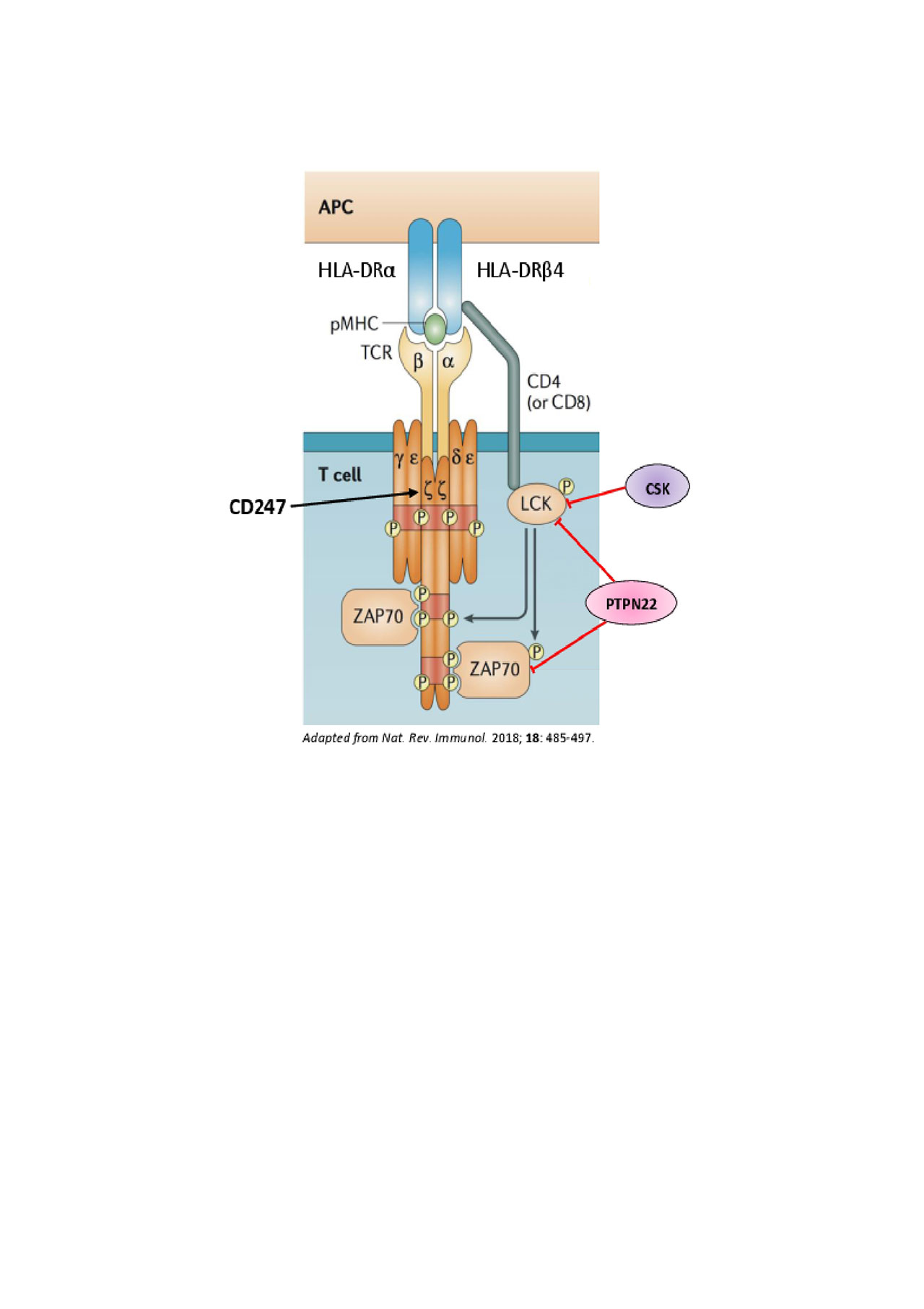
Conclusion: This is the largest genetic study of African American SSc patients identifying classical HLA alleles increasing risk in clinical subsets of SSc. Class II HLA alleles are responsible for increasing risk for scleroderma in general and the class I HLA alleles play a more prominent role in defining the extent of skin involvement. Increased risk from the HLA genes along with previously implicated genes in SSc susceptibility (CD247, CSK, PTPN22) together with the newly identified role of HLA-DRB4 point towards the important role of CD4 T cells and the relevance of T cell receptor (TCR) signaling in SSc pathogenesis.
To cite this abstract in AMA style:
Gourh P, Safran S, Boyden S, Shah A, Mayes M, Doumatey A, Bentley A, Shriner D, Domsic R, Medsger Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Schiopu E, Khanna D, Gordon J, Kron B, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Kolstad K, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Steinbach P, Chandrasekharappa S, Mullikin J, Adeyemo A, Rotimi C, Wigley F, Kastner D, Boin F, Remmers E, Alexander T. Ancestry-Specific Classical HLA Alleles Define Phenotypic Subsets in the African American Scleroderma Population [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/ancestry-specific-classical-hla-alleles-define-phenotypic-subsets-in-the-african-american-scleroderma-population/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/ancestry-specific-classical-hla-alleles-define-phenotypic-subsets-in-the-african-american-scleroderma-population/