Session Information
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: African American (AA) patients have a higher prevalence of SSc than European Americans (EA). Adding to this health disparity, AA SSc patients are more likely to present with pulmonary involvement leading to significant morbidity and mortality. Samples from the Genome Research in African American Scleroderma Patients (GRASP) cohort were used for a genome-wide association analysis (GWAS) study in AA SSc patients. Intronic variants within the intraflagellar transport 43 (IFT43) gene were identified as the top non-HLA loci that were significant at the genome-wide level. These variants were in tight linkage disequilibrium with each other and were African-ancestry specific.
Methods: Expression quantitative trait loci (eQTL) analysis of fibroblast data from the Genotype-Tissue Expression (GTEx) project was performed for the IFT43 variants. Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and DNase I hypersensitive sites sequencing (DNase-Seq) sample data from the GEO and ENCODE databases were downloaded and filtered to exclude poor-quality samples. 251 samples from primary cells and tissues, covering 76 distinct primary tissues and cell types, were analyzed further. Real-time PCR (RT-PCR) was performed on primary fibroblast cells with and without the IFT43 risk variants. Two fibroblast cell lines, BJ and IMR-90, were obtained and processed for ATAC-Seq and RNA-Seq.
Results: eQTL analysis of fibroblast data from GTEx revealed increased expression of TGFB3 in samples with the IFT43 minor variants, suggesting that TGFB3 expression was likely being regulated by these IFT43 intronic variants (Figure 1). RT-PCR for TGFB3 expression was performed on fibroblasts cells from healthy AA individuals. Individuals heterozygous for the IFT43 minor variant had 1.6-fold higher TGFB3 expression compared to wildtype individuals (Figure 2). Bioinformatic analysis of ATAC-Seq and DNase-Seq data for fibroblasts showed that one of the SSc-associated IFT43 variants was within an open, accessible chromatin region (Figure 3A). ATAC-Seq analysis of the BJ and IMR-90 fibroblast cell lines confirmed accessible chromatin at the site of one of the IFT43 variants (Figure 3B).
Conclusion: Based on these results, we hypothesize that the IFT43 variants overlap cis-regulatory modules (i.e. enhancer elements) and disrupt transcription factor binding, leading to altered TGFB3 gene expression and function in SSc patients. TGFB3 could be regulating fibrosis via the canonical TGFB pathway or be inducing pathogenic Th17 cell development. Future directions include deletion of these IFT43 cis-regulatory modules in fibroblast cell lines utilizing CRISPR-Cas9 and assessing for changes in TGFB3 gene expression. ChIP-Seq will be used for identifying transcription factor binding at the site of SSc-associated variants. This work implicates TGFB3 as a novel therapeutic target in SSc, especially with regard to regulating fibrosis and autoimmunity. An isoform-selective TGFB inhibitor (blocking TGFB1 and -B3) is in early phase clinical trials in SSc. Identifying SSc patients based on variants increasing TGFB3 expression would provide an ideal target population for such a therapeutic agent.
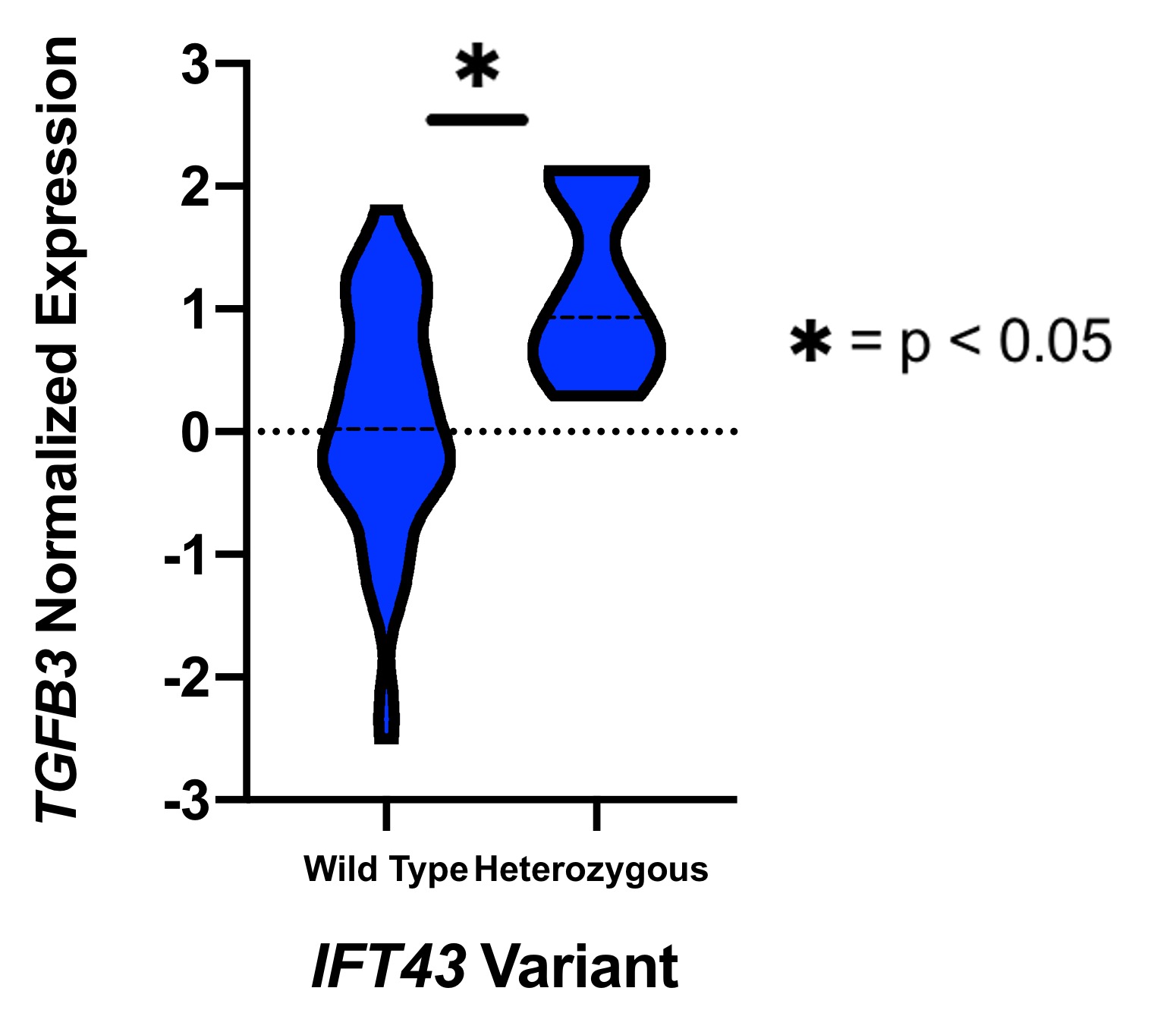 Figure 1. GTEx TGFB3 Expression.
Figure 1. GTEx TGFB3 Expression.
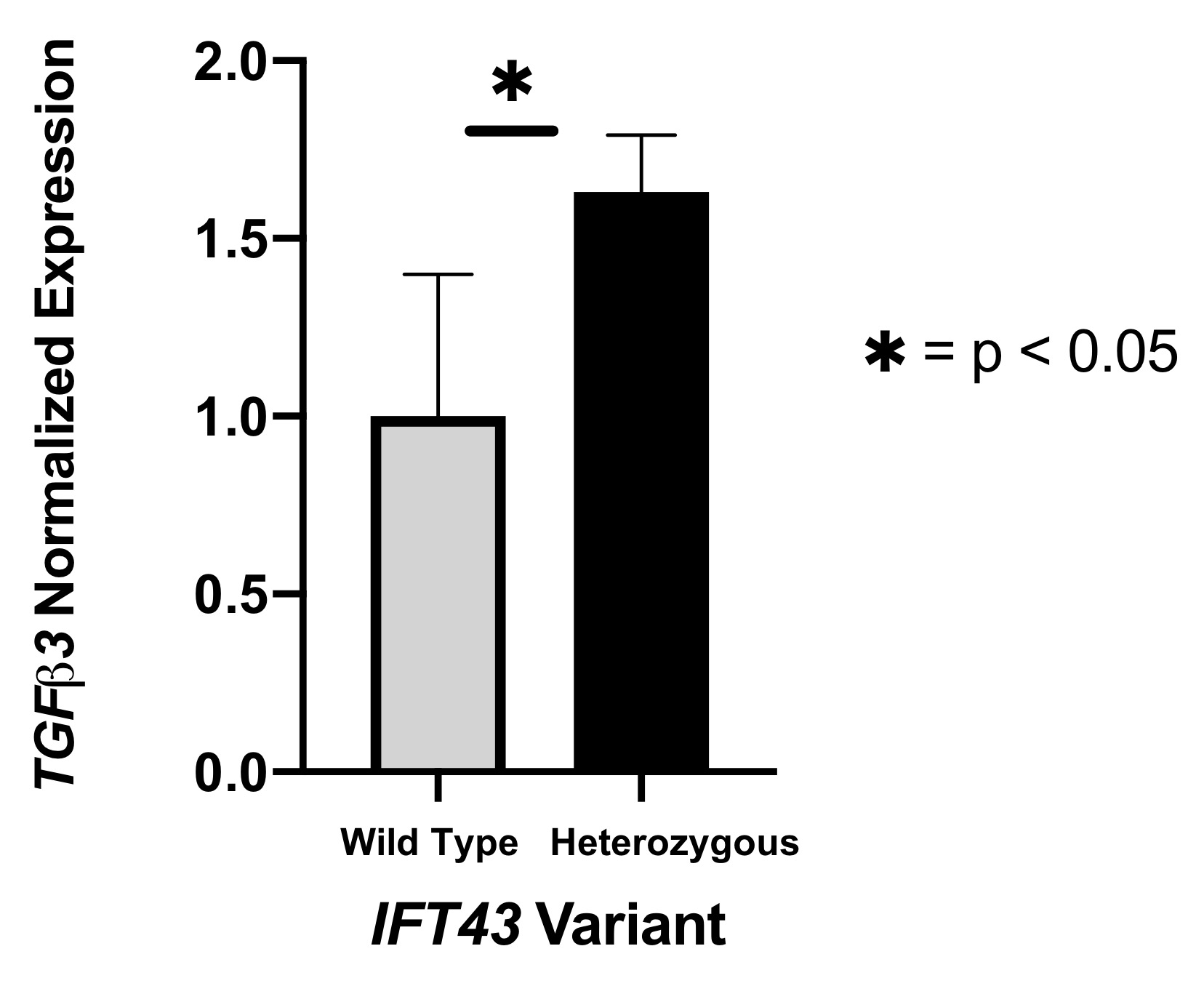 Figure 2. RT-PCR for TGFB3 expression.
Figure 2. RT-PCR for TGFB3 expression.
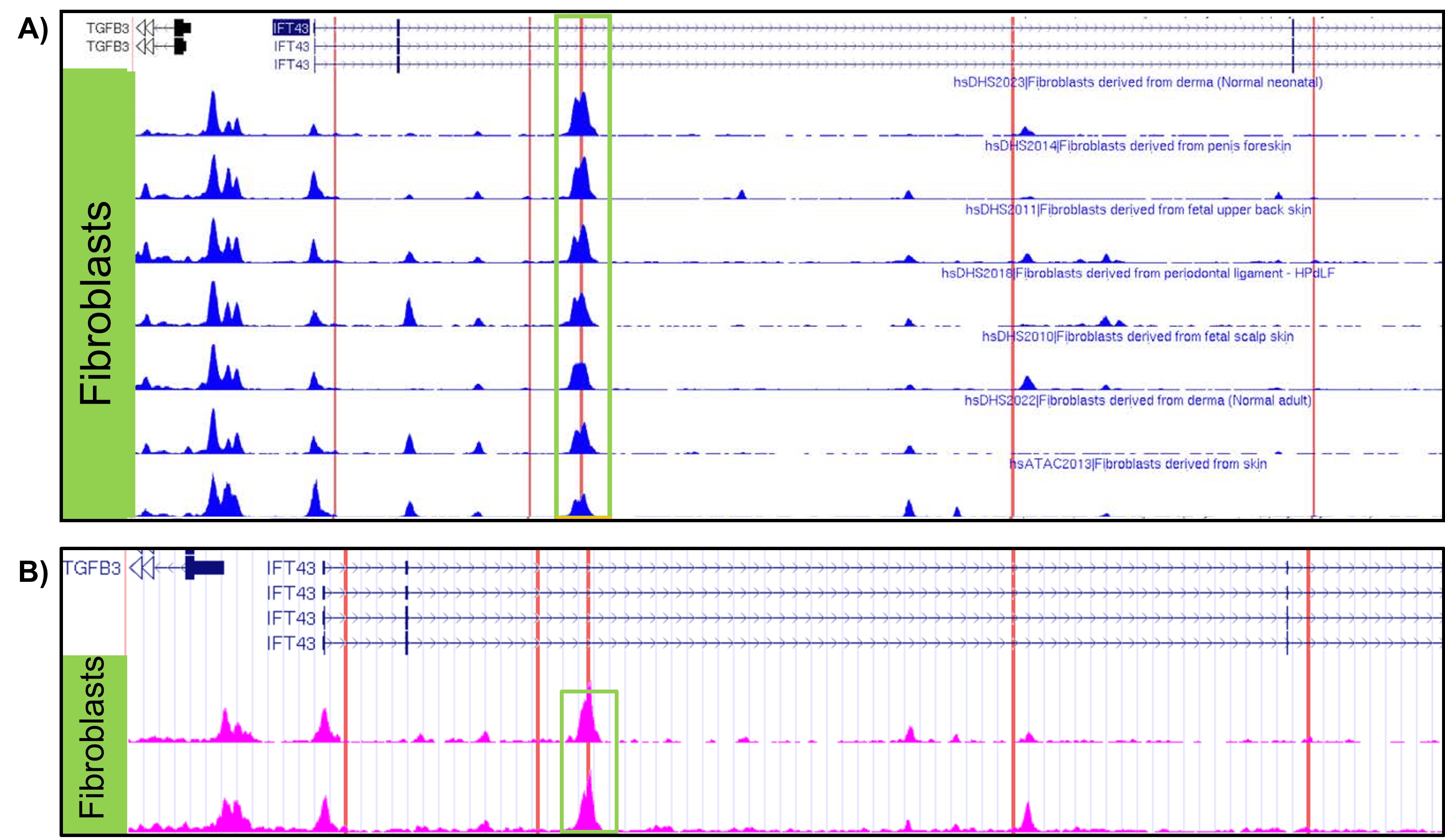 Figure 3. A) ATAC-Seq and DNAse-Seq tracks from ENCODE database showing accessible chromatin in fibroblasts. B) ATAC-Seq results from obtained BJ and IMR-90 fibroblast cell lines. Red lines represent location of IFT43 variants.
Figure 3. A) ATAC-Seq and DNAse-Seq tracks from ENCODE database showing accessible chromatin in fibroblasts. B) ATAC-Seq results from obtained BJ and IMR-90 fibroblast cell lines. Red lines represent location of IFT43 variants.
To cite this abstract in AMA style:
Hartman J, Conte A, Borden C, Kaundal U, Zhao Y, Safran S, Shah A, Mayes M, Doumatey A, Bentley A, Shriner D, Domsic R, Medsger T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Schiopu E, Khanna D, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Kolstad K, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Hinchcliff M, Chandrasekharappa S, Dell'Orso S, Adeyemo A, Rotimi C, Remmers E, Wigley F, Kastner D, Boin F, Casellas R, Gourh P. African Ancestry-Specific Variants Regulate TGFB3 Expression in Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/african-ancestry-specific-variants-regulate-tgfb3-expression-in-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/african-ancestry-specific-variants-regulate-tgfb3-expression-in-systemic-sclerosis/
