Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: There are significant racial disparities in systemic sclerosis (SSc), with an increased disease burden and worse outcomes among African American (AA) individuals. Reasons for this increased prevalence, severity, and mortality of SSc in AA patients are largely unknown. AA individuals primarily derive their ancestry from West Africa. AA SSc patients, like West Africans, have a higher prevalence of SSc with an earlier age of onset, diffuse skin involvement and pulmonary complications. The objective of this study is to identify genetic variants that explain the increased frequency and severity of SSc in AAs.
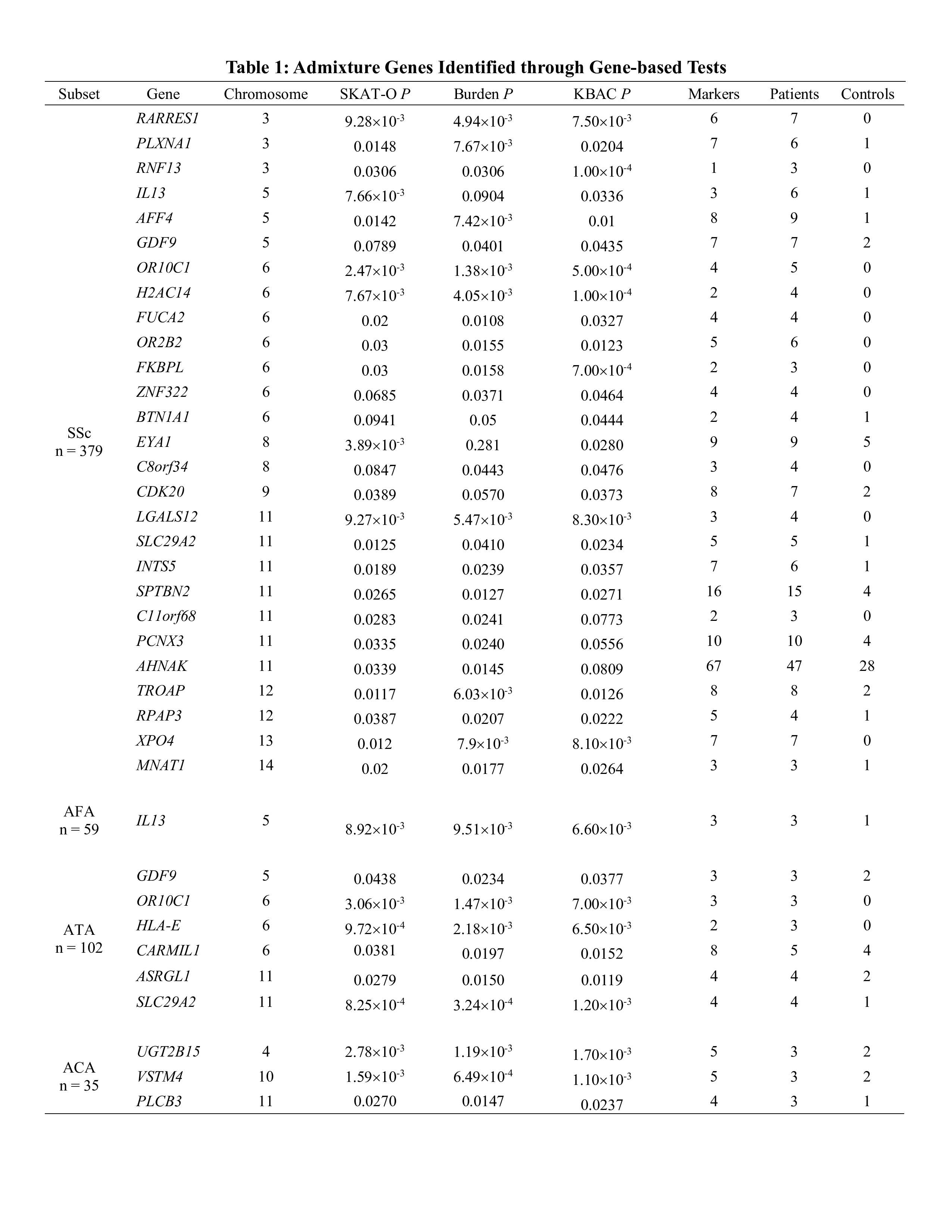
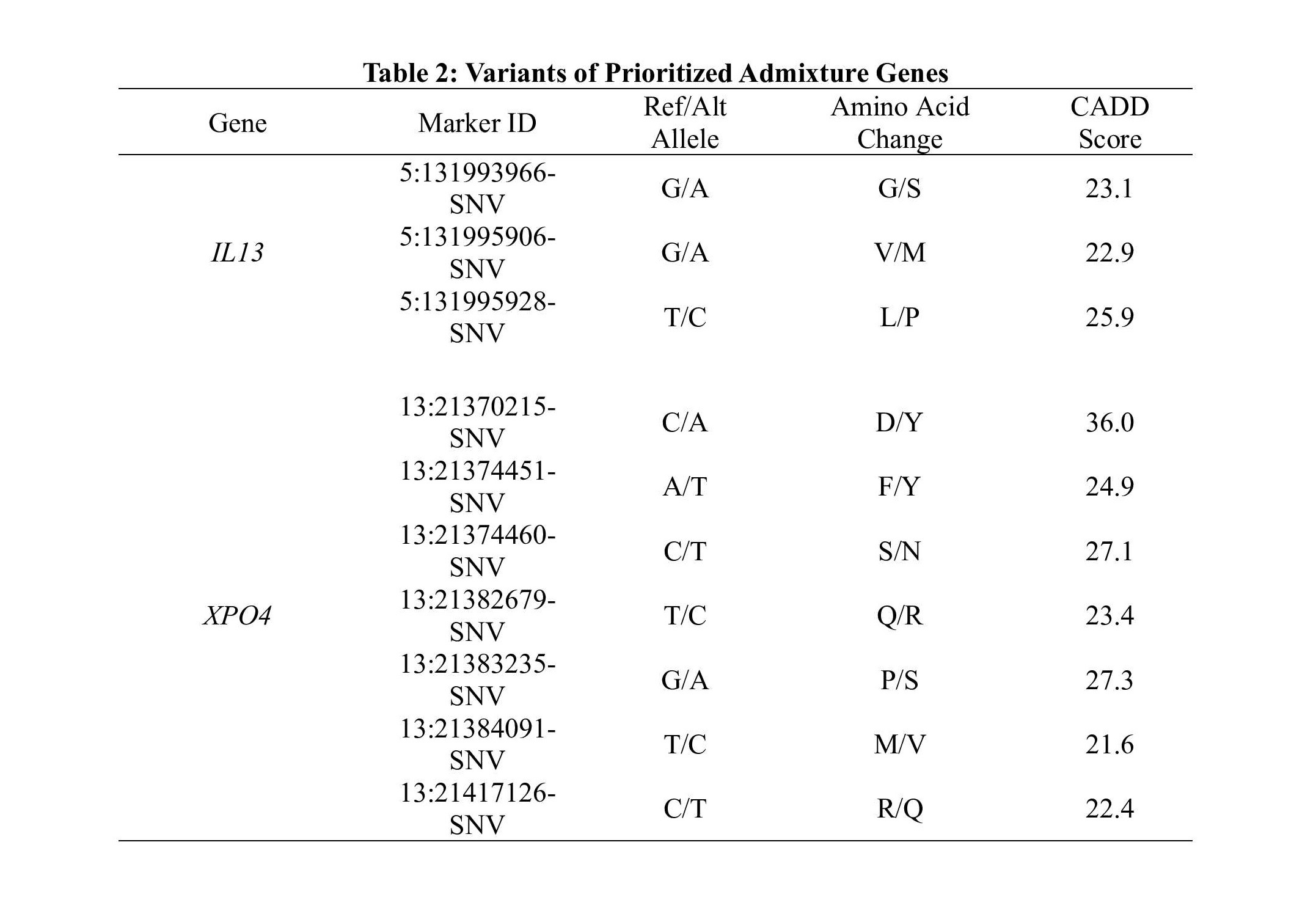
Methods: SSc patients from the Genome Research in African American Scleroderma Patients (GRASP) cohort met either the 1980 ACR or 2013 EULAR criteria for SSc, or had at least 3 of 5 features of CREST syndrome. Admixture mapping was performed on 946 patients and 934 controls using 998,750 genotyped markers. Local ancestry was inferred using RFMix (v.1.5.4) and a reference panel of five ancestries derived from the 1000 Genomes Project. The genome-wide significance level was set at 2.3×10-4 and suggestive level was P< 0.05. Genes contained within the admixed regions with at least nominal significance were identified. Gene-based (SKAT-O, Burden, and KBAC) testing was performed on missense or loss of function (LoF) variants with a CADD score >10 suggesting deleteriousness and a minor allele frequency < 0.001 in gnomAD and TOPMed, from the exome sequences of 379 patients and 411 controls. Genes with a P< 0.05 in at least two of three gene-based tests were identified for further analysis.
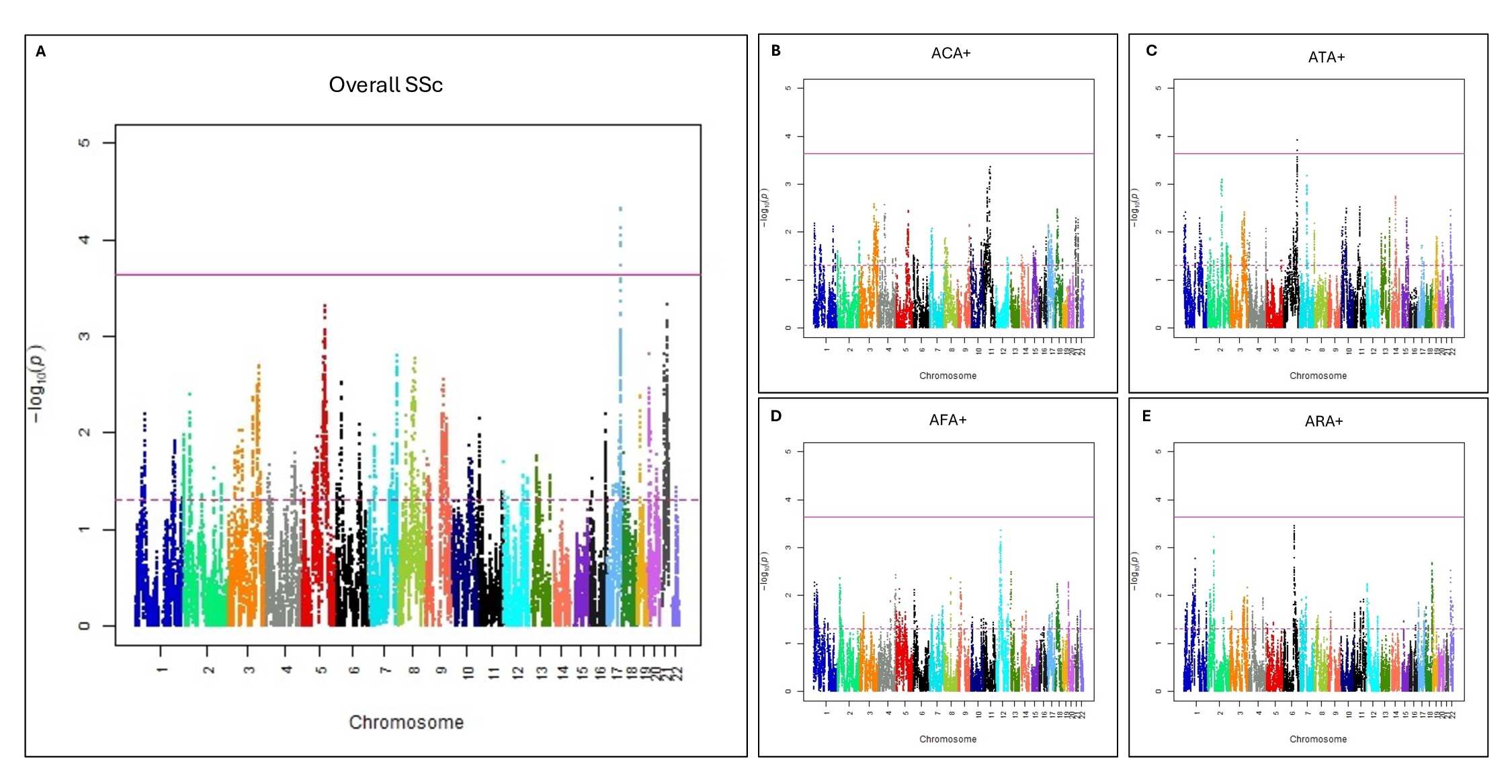
Results: Admixture mapping of SSc identified one genome-wide significant and thirteen suggestive regions (Figure 1A). Stratified analysis on SSc autoantibody subsets of SSc identified one genome-wide significant and several suggestive regions (Figure 1B-E). Gene-based testing within these admixed regions identified enrichment of rare variants in twenty-seven genes in overall SSc and one, six, and three genes in the AFA, ATA, and ACA subsets, respectively (Table 1). Four genes (IL13, SCL29A2, OR10C1, and GDF9) were significant in both overall SSc and an autoantibody subset.
Conclusion: Our unique approach of utilizing admixture mapping to map trait gene regions and analyze them for enrichment of rare variants has yielded several potential candidate genes. Perhaps most easily understood are IL13, GDF9, and XPO4, which regulate TGFβ signaling via SMAD3. IL13 encodes an immunoregulatory cytokine that serves to activate an immune response through inflammation and activation of the pro-fibrotic TGFβ pathway. GDF9 encodes a secreted ligand for TGFβ receptors, initiating recruitment and activation of SMAD3. XPO4 is associated with anti-fibrotic activity via mediation of the nuclear export of SMAD3 and regulation of TGFβ signaling. Dysregulated IL13 and TGFβ signaling have been previously reported in SSc. Our unique approach and these findings help explain some of the genetic risk factors in AA subjects with SSc that contribute to the disparate outcomes in this population.
To cite this abstract in AMA style:
Hicks J, Shriner D, Shah A, Mayes M, Doumatey A, Bentley A, Domsic R, Medsger, Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Khanna D, Schiopu E, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Thomas J, Remmers E, Adeyemo A, Rotimi C, Wigley F, Boin F, Kastner D, Gourh P. Admixture Mapping and Gene-Based Analysis Identifies Rare Variants in Genes in the IL-13 and TGFβ Signaling Pathways in African Americans with Systemic Sclerosis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/admixture-mapping-and-gene-based-analysis-identifies-rare-variants-in-genes-in-the-il-13-and-tgf%ce%b2-signaling-pathways-in-african-americans-with-systemic-sclerosis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/admixture-mapping-and-gene-based-analysis-identifies-rare-variants-in-genes-in-the-il-13-and-tgf%ce%b2-signaling-pathways-in-african-americans-with-systemic-sclerosis/