Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Knee osteoarthritis (KOA) is a heterogeneous condition characterized by changes in a variety of joint tissues and driven by a number of different potential mechanisms. The purpose of this study was to explore machine learning approaches to phenotyping in KOA in order to better define the progression phenotype(s) that may be more responsive to interventions.
Methods: We focused on identifying baseline differences in the Johnston County OA Project (T1) between 1) knees with and without prevalent radiographic KOA (rKOA, Kellgren-Lawrence grade [KLG] 2 or more, “OA”), and 2) those that did or did not have worsening KLG and/or symptoms (progression) at follow-up. The dataset included observations on 741 participants with 78 baseline variables selected to represent clinically relevant characteristics of this sample. Both k-means and hierarchical clustering methods were applied to identify important subgroups of observations within the dataset. Two high-dimension low-sample size (HDLSS) methods were used: SigClust, to assess statistical significance of clusters and 2) DiProPerm, a projection and permutation-based approach, to compare groups (e.g., prevalent rKOA vs. no OA, progression vs. non-progression) by testing equality of distributions (by z-score) over all variables. Given multiple comparisons, a z-score of at least 2 (p value<0.05) was considered statistically significant.
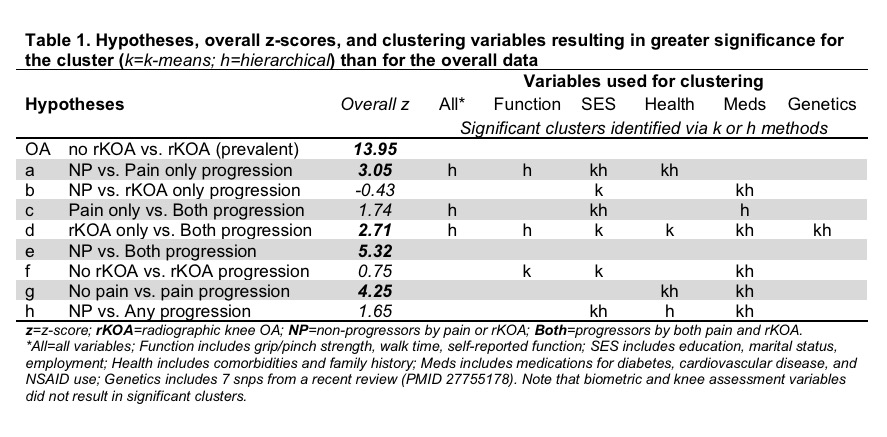
Results: When considering all observations and all variables simultaneously, significant differences were observed for 5 comparisons (Table 1, hypotheses “OA”, a, d, e, and g). Both k-means and hierarchical clustering methods identified clusters with greater significance than using all the observations, suggesting the existence of groups of observations where the difference between progressors and non-progressors is enhanced. For example, when focusing on the genetic data, both clustering methods identified different clusters of observations using SigClust (p<0.01), and further showed that when observations were clustered according to the genetic variables, there were stronger results for hypothesis d (rKOA only vs. both radiographic and symptomatic progression). Statistically significant clusters were also identified when clustering was based on physical function, socioeconomic status, health, and medication variables. Clustering on all variables resulted in significant clusters only under hierarchical methods.
Conclusion: These innovative methods provide a way to assess numerous variables of different types and scalings simultaneously in relation to KOA prevalence and progression and could be used for assessing other outcomes of interest. Such methodology could identify both known and novel KOA phenotypes, potentially improving patient selection for specific interventions and providing insight into pathophysiology of this heterogeneous condition.
To cite this abstract in AMA style:
Nelson A, Fuller M, Cleveland B, Schwartz T, Jordan JM, Callahan LF, Marron JS, Loeser R. A Machine Learning Approach to Knee OA Phenotyping: The Johnston County Osteoarthritis Project [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/a-machine-learning-approach-to-knee-oa-phenotyping-the-johnston-county-osteoarthritis-project/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/a-machine-learning-approach-to-knee-oa-phenotyping-the-johnston-county-osteoarthritis-project/