Session Information
Session Type: Abstract Session
Session Time: 11:45AM-12:00PM
Background/Purpose: The use of IL-23 inhibitors (IL-23i) for psoriatic diseases resulted in significant improvement in disease symptoms for many patients. Whether any blood immune cell profiles can predict clinical response to IL-23 blockade has not been examined, however. Here, we applied single cell RNA-seq immune profiling to blood samples from 27 PsO/A patients to identify cellular features associated with response to IL-23 blockade by tildrakizumab in the setting of a clinical trial.
Methods: PBMCs from psoriasis (n = 21) and psoriatic arthritis (n=6) patients were collected as part of the MINIMA study (NCT04271540) at Brigham and Women’s Hospital before (following washout) and after 24 weeks of treatment with tildrakizumab. Patients currently had moderate or severe PsO. PASI scores were recorded before and after treatment, and % improvement of 50% or 70% was calculated. 48% of participants had a PASI improvement of 70% or greater, and 78% PASI improvement of 50% or greater. Table 1 denotes the disease characteristics and demographics of the patients. Single-cell RNA sequencing was conducted on baseline samples before treatment and on follow-up samples, followed by QC, batch correction, and unsupervised clustering. Analyses were performed using Wilcoxon paired tests, covarying neighborhood analysis (CNA), mixed effects association of single cell analysis (MASC), and differential gene expression analysis.
Results: We compared baseline samples from patients with greater or less than 70% improvement to identify baseline immune features associated with response. Assessment of PBMCs at a broad level did not reveal associations of any major immune cell populations with better or worse response to IL-23i. We then focused on T cell populations likely to be affected by IL-23i especially IL-23R+ T cells (Fig 1). The IL23R+ CD4 T cell had phenotypes consistent with Th17 and Th1 cells, including expression of IFNG, CXCR3, and IL17A, CCR6 and RORC. As a whole, the proportion of these cells in blood did not change following IL-23i. Further subcluster analysis of the IL23R+ CD4 T cell clusters revealed an association of a subcluster of cells with 70% clinical improvement and a separate subcluster of cells associated with lesser clinical improvement (Fig 2). Response-associated CD4+ T cells had a gene signature which corresponded to pathogenic Th17 cells (Hu et al, Nat. Commun. 2017). Additionally, patients with improvement >50% had higher expression of CCL5 and STAT1 in the IL23R+ CD4 T cells. To understand relevance of these cells to cells that are expanded in psoriasis skin, we applied a gene module score of genes associated with IFNg+ IL-17A+ cells from psoriasis skin (Kim et al, J. Allergy Clin. Immunol., 2021). This gene score was also higher in the response-associated cluster.
Conclusion: We have identified a subset of CD4+ T cells whose abundance in circulation before treatment is associated with response to IL-23i in skin disease severity. This T cell population demonstrates transcriptomic features that correspond to those expanded in psoriasis lesional skin. These results suggest that cellular features detectable in blood may be informative in predicting clinical response to IL-23 blockade.
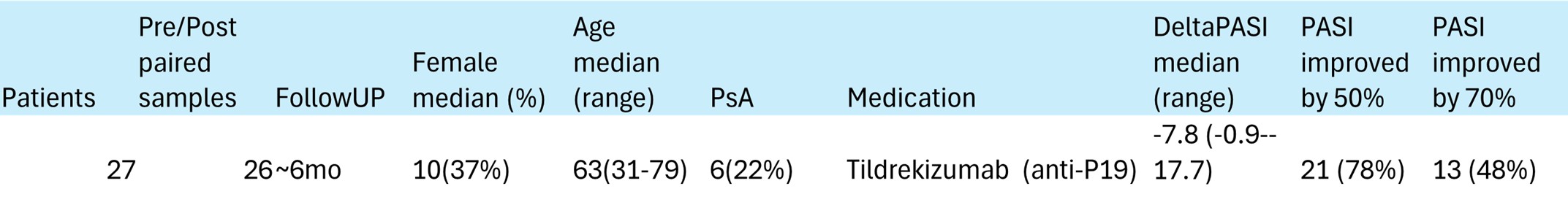 Table 1. Description of patients included in the scRNAseq from the MINIMA trial.
Table 1. Description of patients included in the scRNAseq from the MINIMA trial.
.jpg) Figure 1. A) scRNAseq of PBMCs from IL-23i-treated patients, including cells from baseline (pre-treatment) and week 24 post-treatment samples (~136K cells). B) Mixed effects association of single cell analysis demonstrates a lack of association of PBMC clusters with pre- or post-IL-23i. C) CD4 T cells subset from PBMCs. D) IL23R, IL17A, and IFNexpression in CD4 T cells.
Figure 1. A) scRNAseq of PBMCs from IL-23i-treated patients, including cells from baseline (pre-treatment) and week 24 post-treatment samples (~136K cells). B) Mixed effects association of single cell analysis demonstrates a lack of association of PBMC clusters with pre- or post-IL-23i. C) CD4 T cells subset from PBMCs. D) IL23R, IL17A, and IFNexpression in CD4 T cells.
.jpg) Figure 2. A) Subcluster of CD4+IL23R+ clusters. B) MASC analysis of pre-treatment samples demonstrating CD4+IL23R+ subclusters associated with baseline (pre-treatment) or week 24 (post-treatment). C) Module scores of genes that predict Th17 cell pathogenicity and genes highly expressed in psoriasis lesional skin. Both scores are higher in the better improvement-associated cluster than the lesser improvement-associated cluster. D) Mean expression of CCL5 and STAT1 in pre-treatment samples is higher in those who have greater than 50% improvement from IL-23i at week 24.
Figure 2. A) Subcluster of CD4+IL23R+ clusters. B) MASC analysis of pre-treatment samples demonstrating CD4+IL23R+ subclusters associated with baseline (pre-treatment) or week 24 (post-treatment). C) Module scores of genes that predict Th17 cell pathogenicity and genes highly expressed in psoriasis lesional skin. Both scores are higher in the better improvement-associated cluster than the lesser improvement-associated cluster. D) Mean expression of CCL5 and STAT1 in pre-treatment samples is higher in those who have greater than 50% improvement from IL-23i at week 24.
To cite this abstract in AMA style:
Marks K, Sun A, Adejoorin I, Perez Chada L, Perillo A, Barrett L, Garshick M, Krueger J, Merola J, DiCarli M, Rao D, Weber B. Single cell RNA-seq immune profiling of blood samples from a psoriatic disease clinical trial reveals a CD4 T cell population associated with response to IL-23 blockade [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/single-cell-rna-seq-immune-profiling-of-blood-samples-from-a-psoriatic-disease-clinical-trial-reveals-a-cd4-t-cell-population-associated-with-response-to-il-23-blockade/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-seq-immune-profiling-of-blood-samples-from-a-psoriatic-disease-clinical-trial-reveals-a-cd4-t-cell-population-associated-with-response-to-il-23-blockade/
