Session Information
Date: Wednesday, October 29, 2025
Title: Abstracts: Rheumatoid Arthritis – Treatment II: Phenotyping and Personalization (2639–2644)
Session Type: Abstract Session
Session Time: 9:30AM-9:45AM
Background/Purpose: Oral methotrexate (MTX) is the cornerstone treatment for newly diagnosed rheumatoid arthritis (RA), yet up to 50% of patients do not respond adequately. Early prediction of MTX response is crucial to prevent disease progression and promptly initiate alternative therapies. Recent studies suggest a link between the gut microbiome and MTX efficacy in treatment-naïve RA patients. However, the use of 16S rRNA sequencing in these studies limits taxonomic resolution in identifying gut microbial relationships with treatment response; and the response criteria applied were arbitrary, as acknowledged by the investigators. To enhance clinical applicability, our study used whole genome shotgun metagenome sequencing and the standardized EULAR response criteria to rigorously investigate associations between pre-treatment gut microbiome and MTX response.
Methods: A cohort of 60 treatment-naïve RA patients (mean age 51.2 years) initiated treatment with oral MTX at a low dose (15 mg/week). Of these, 43 were female, 6 were current smokers, and 14 were on prednisone (Table 1). Stool samples collected at baseline and after 3–3.5 months of treatment underwent metagenomic sequencing. Patients were classified as MTX responders (n = 37) or non-responders (n = 23) based on the EULAR response criteria. Response was defined as a decrease in DAS28-CRP (ΔDAS28-CRP) > 1.2, or ΔDAS28-CRP > 0.6 with a follow-up DAS28-CRP ≤ 5.1. Differential abundance and prevalence analyses identified microbiome features distinguishing the two groups. A gradient-boosting linear regression model using baseline gut microbiome and clinical data (BMI, DAS28-CRP, ACPA status, and prednisone dose) was developed to predict MTX response.
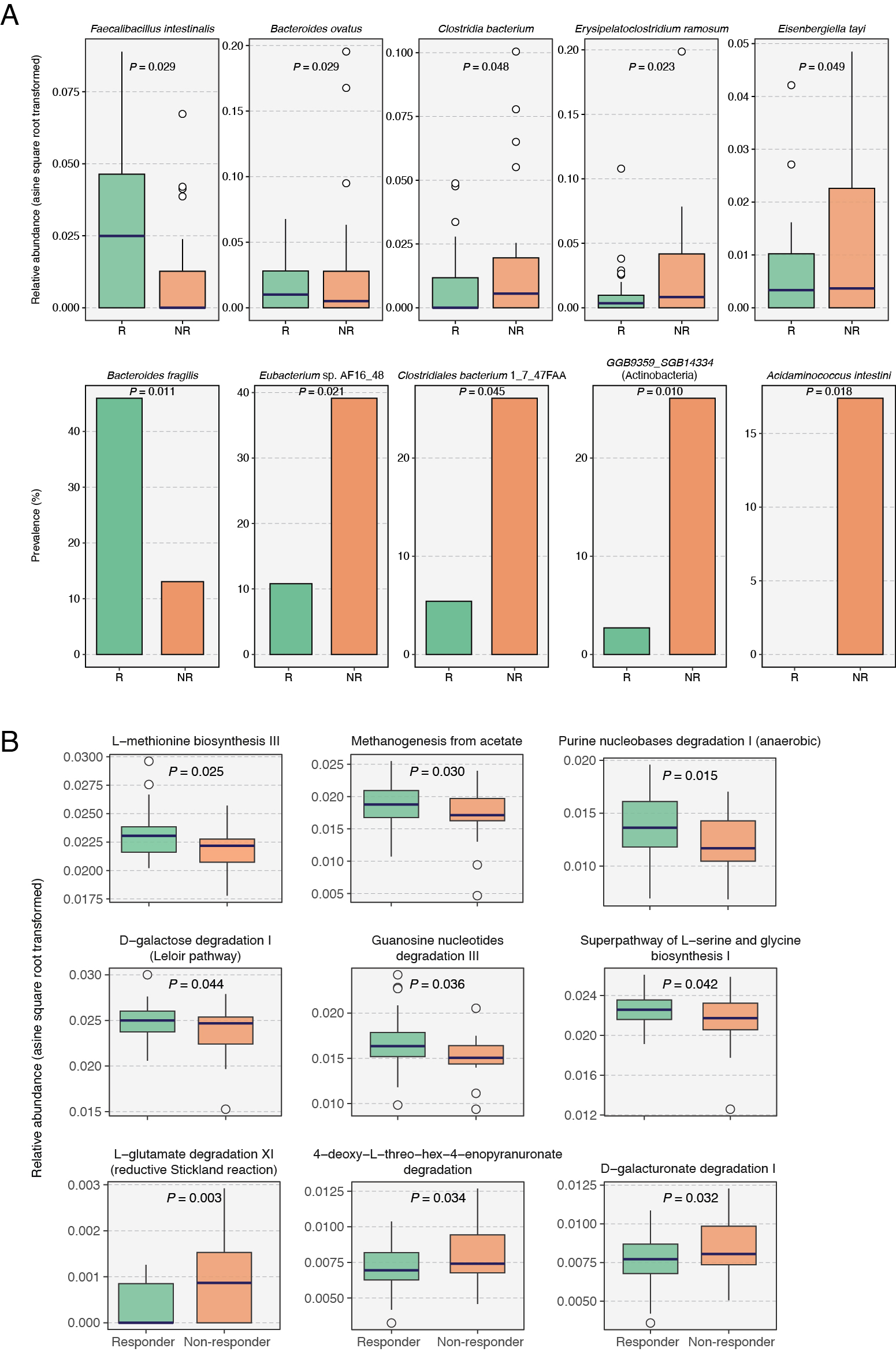
Results: Baseline microbiome analysis revealed significant differences between responders and non-responders, with five species and nine metabolic pathways having differential abundance (Fig. 1). Additionally, clinical response correlated strongly with the presence of specific species, such as Bacteroides fragilis and Eubacterium sp. AF16-48 (Fig. 1). Additionally, the odds of MTX response increased ten-fold in patients harboring any of the top ten differentially prevalent gut microbial species (OR = 10.4). Moreover, genes of the dihydrofolate reductase (DHFR) enzyme were significantly less abundant in responders (P = 0.031). Finally, our machine learning model using baseline gut microbiome and clinical data demonstrated an AUC of 0.83 in leave-one-out cross-validation, outperforming an AUC of 0.72 from clinical data alone. Our model achieved an AUC of 0.77 in an external validation cohort of 38 treatment-naïve RA patients (Fig. 2).
Conclusion: We identified gut microbiome features associated with MTX response in early RA patients. Our machine learning model suggests the potential of gut microbiome profiling as a personalized predictive tool for MTX outcomes.
.jpg) Figure 1. Differences in baseline gut microbial species (A) and MetaCyc pathways (B) between responders and non-responders. Multiple linear regression models adjusted for age and BMI were used to test for the statistical significance to identify the differentially abundant microbiome features between both patient groups. Differentially prevalent species were identified using Fisher’s exact test. R, Responder; NR, Non-responder.
Figure 1. Differences in baseline gut microbial species (A) and MetaCyc pathways (B) between responders and non-responders. Multiple linear regression models adjusted for age and BMI were used to test for the statistical significance to identify the differentially abundant microbiome features between both patient groups. Differentially prevalent species were identified using Fisher’s exact test. R, Responder; NR, Non-responder.
.jpg) Figure 2. Performance of microbiome-based gradient-boosting linear regression model in predicting the MTX response. (A) Feature importance of microbial species that were used in the model. (B) Scatter plot between predicted ΔDAS28CRP and actual ΔDAS28CRP values. (C) Leave-one-out cross-validation receiver operating characteristic (ROC) curve of the gradient boosting machine-learning model predicting response to MTX from the training dataset. (D) ROC curve of the gradient boosting machine-learning model predicting response to MTX from an external validation cohort.
Figure 2. Performance of microbiome-based gradient-boosting linear regression model in predicting the MTX response. (A) Feature importance of microbial species that were used in the model. (B) Scatter plot between predicted ΔDAS28CRP and actual ΔDAS28CRP values. (C) Leave-one-out cross-validation receiver operating characteristic (ROC) curve of the gradient boosting machine-learning model predicting response to MTX from the training dataset. (D) ROC curve of the gradient boosting machine-learning model predicting response to MTX from an external validation cohort.
To cite this abstract in AMA style:
GUPTA V, Koller A, Hur B, Bailey M, Delger K, myasoedova E, Kronzer V, Davis J, Sung J. Gut Microbiome Signatures Forecast Clinical Response to Methotrexate in Treatment-Naïve Early Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/gut-microbiome-signatures-forecast-clinical-response-to-methotrexate-in-treatment-naive-early-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gut-microbiome-signatures-forecast-clinical-response-to-methotrexate-in-treatment-naive-early-rheumatoid-arthritis/