Session Information
Session Type: Abstract Session
Session Time: 3:15PM-3:30PM
Background/Purpose: The oral microbiome plays a critical role in modulating systemic inflammation and maintaining immune homeostasis, partly through its interactions with the gut microbiome. Although gut microbiome dysbiosis has been implicated in symptomatic hand osteoarthritis (SHOA), the role of oral microbiome dysbiosis in SHOA and its potential correlation with gut microbiome remains unclear. Elucidation these relationships may provide novel insights into the pathogenesis of SHOA and inform future treatment strategies.
Methods: Participants were recruited from the Xiangya Osteoarthritis (OA) Study, a community-based cohort study. SHOA was defined as the presence of both self-reported pain and radiographic OA in at least one hand. The control group comprised participants without hand pain or radiographic OA. Among the 764 participants included in this study, 52 (6.8%) had SHOA. Saliva samples were analyzed using 16S rRNA gene sequencing. Oral microbial richness (α-diversity, measured by Shannon index) and composition (β-diversity, measured by Bray-Curtis distance) were compared between SHOA and control groups. Associations of the relative abundance of oral microbiome genera with SHOA and its severity were examined. Functional alterations in the oral microbiome were inferred using Kyoto Encyclopedia of Genes and Genomes (KEGG) level 3 pathways analysis. Gut microbiome data from a previous study conducted within the same cohort were integrated to examine ecological links between the oral and gut microbiomes.1 Correlations between SHOA-associated oral microbial genera and SHOA-associated gut microbial functions were also assessed. A workflow is presented in Fig. 1.
Results: Compared with controls, participants with SHOA exhibited significantly lower oral microbial α-diversity (P=0.007, Fig. 2A) and altered β-diversity (P=0.007, Fig. 2B). A total of 911 amplicon sequence variants and eight genera were uniquely identified in the SHOA group (Fig. 2C). In both the SHOA and control groups, the oral microbiome was predominantly composed of the genera Neisseria, Streptococcus, Haemophilus, Porphyromonas, Fusobacterium(Fig. 2D). The relative abundance of the genus Trichococcus was significantly higher in SHOA participants (P=0.001, Q=0.073, Fig. 2E-F), and was positively correlated with SHOA severity. Oral microbiome KEGG level 3 pathways involved in tropane, piperidine, and pyridine alkaloid biosynthesis (P=0.006, Q=0.084), as well as indole alkaloid biosynthesis (P=0.006, Q=0.090), were significantly inversely associated with SHOA. Furthermore, the number of correlations in the oral-gut microbiome network was significantly reduced in SHOA participants compared with controls (Fig. 3A). Additionally, a positive correlation was observed between oral microbiota Trichococcus and the gut microbiome KEGG pathway of tyrosine metabolism, both of which were significantly associated with SHOA (P=0.001, Q=0.047, Fig. 3B-C).
Conclusion: Oral microbiome dysbiosis and the disrupted oral-gut microbiome network are associated with prevalent SHOA, suggesting a potential role for oral-gut microbiome interactions in disease pathogenesis.REFERENCES1. Wei J, et al. EBioMedicine 2023;98:104892.
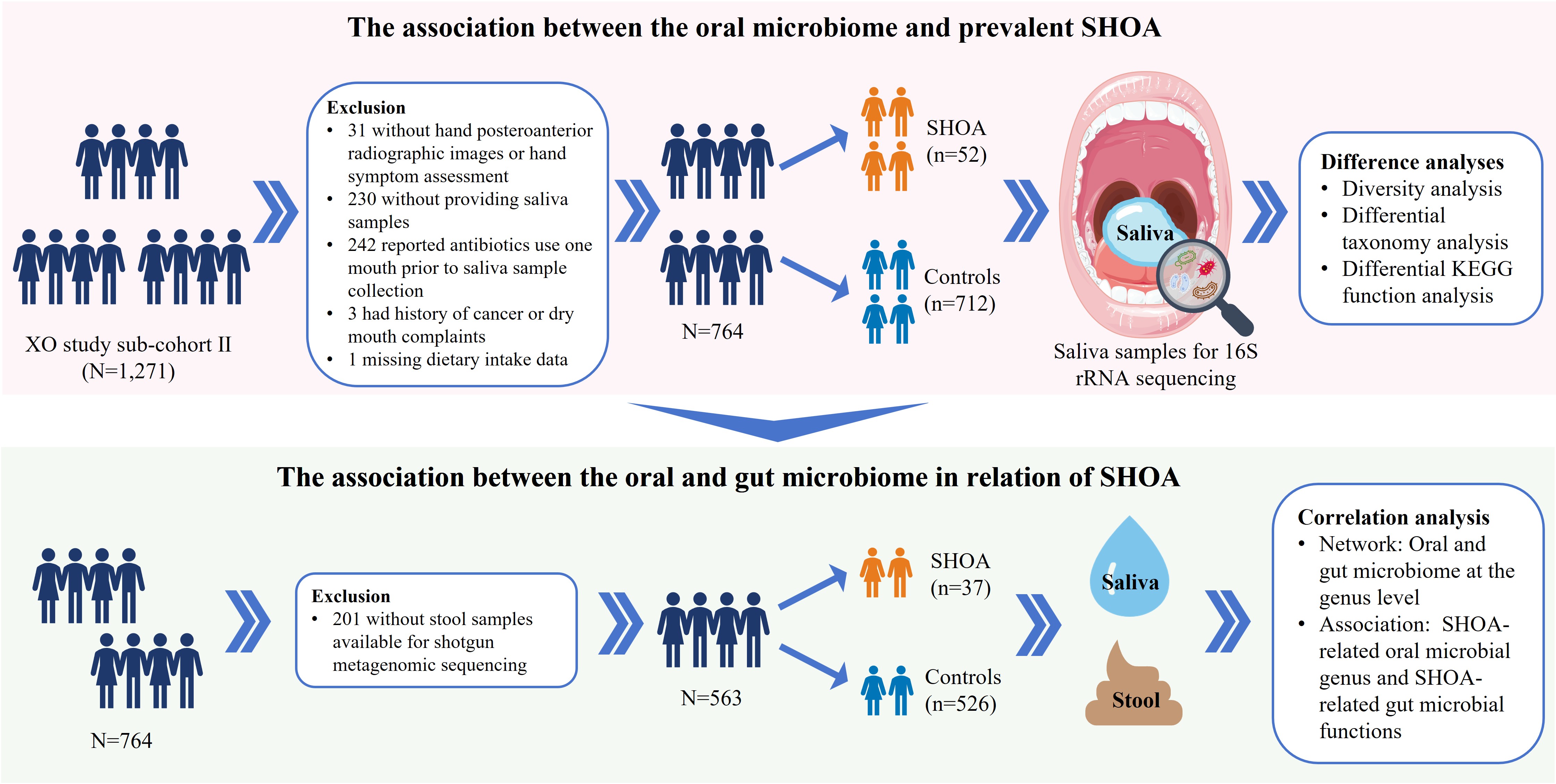 Figure 1. Summary of the present study. SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 1. Summary of the present study. SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
.jpg) Figure 2. Comparison of oral microbiome diversity and composition between participants with SHOA and controls. (A) Violin plots showing α-diversity measured by the Shannon index between participants with SHOA and controls. (B) PCoA plots comparing β-diversity measured by Bray-Curtis distance between participants with SHOA and controls. (C) Venn diagram illustrating shared and exclusive ASVs and microbial genera between participants with SHOA and controls. (D) Relative abundance of the top 20 microbial genera in the oral microbiome composition of participants with SHOA and controls. (E) Volcano plot depicting differences in the microbial genera between participants with SHOA and controls. The red region indicates statistically significant differences (Q < 0.1). (F) Relative abundance of differential microbial genus between participants with SHOA and controls. SHOA, symptomatic hand osteoarthritis; PCoA, principal coordinates analysis; ASVs, amplicon sequence variants.
Figure 2. Comparison of oral microbiome diversity and composition between participants with SHOA and controls. (A) Violin plots showing α-diversity measured by the Shannon index between participants with SHOA and controls. (B) PCoA plots comparing β-diversity measured by Bray-Curtis distance between participants with SHOA and controls. (C) Venn diagram illustrating shared and exclusive ASVs and microbial genera between participants with SHOA and controls. (D) Relative abundance of the top 20 microbial genera in the oral microbiome composition of participants with SHOA and controls. (E) Volcano plot depicting differences in the microbial genera between participants with SHOA and controls. The red region indicates statistically significant differences (Q < 0.1). (F) Relative abundance of differential microbial genus between participants with SHOA and controls. SHOA, symptomatic hand osteoarthritis; PCoA, principal coordinates analysis; ASVs, amplicon sequence variants.
.jpg) Figure 3. Oral-gut microbiome correlation networks at the genus level in SHOA group and control group and the association between SHOA-related oral microbial genus and SHOA-related gut microbial functions. (A) Only genera with a relative abundance >0.01 and present in >70% of samples were considered. Solid lines indicate significant correlations (P < 0.05) between oral and gut microbial genera, with line width indicating correlation magnitude. Red and blue edges represent positive and inverse correlations, respectively. Red triangles (circles) denote oral (gut) microbial genera with significant differences in relative abundance between the SHOA group and the control group. Yellow triangles (circles) represent oral (gut) microbial genera with no significant differences in relative abundance between the two groups. (B) Heatmap-circle showing correlations between SHOA-related oral microbial genus (i.e., Trichococcus) and SHOA-related gut microbial KEGG pathways. (C) Scatter plot illustrating the correlation between the SHOA-related oral microbial genus (i.e., Trichococcus) and the SHOA-related KEGG pathway (i.e., tyrosine metabolism), which was identified as a significant association. SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 3. Oral-gut microbiome correlation networks at the genus level in SHOA group and control group and the association between SHOA-related oral microbial genus and SHOA-related gut microbial functions. (A) Only genera with a relative abundance >0.01 and present in >70% of samples were considered. Solid lines indicate significant correlations (P < 0.05) between oral and gut microbial genera, with line width indicating correlation magnitude. Red and blue edges represent positive and inverse correlations, respectively. Red triangles (circles) denote oral (gut) microbial genera with significant differences in relative abundance between the SHOA group and the control group. Yellow triangles (circles) represent oral (gut) microbial genera with no significant differences in relative abundance between the two groups. (B) Heatmap-circle showing correlations between SHOA-related oral microbial genus (i.e., Trichococcus) and SHOA-related gut microbial KEGG pathways. (C) Scatter plot illustrating the correlation between the SHOA-related oral microbial genus (i.e., Trichococcus) and the SHOA-related KEGG pathway (i.e., tyrosine metabolism), which was identified as a significant association. SHOA, symptomatic hand osteoarthritis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To cite this abstract in AMA style:
Li J, Xiao Y, Yang T, Hunter d, Zhang W, Doherty M, Zhang Y, Yang Z, Wang Y, Xie D, Li C, Li W, Wen Z, Zeng C, Lei G, Wei J. Oral Microbiome Dysbiosis and Oral-gut Microbial Network Disruption in Hand Osteoarthritis: Data from the Xiangya Osteoarthritis Study [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/oral-microbiome-dysbiosis-and-oral-gut-microbial-network-disruption-in-hand-osteoarthritis-data-from-the-xiangya-osteoarthritis-study/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/oral-microbiome-dysbiosis-and-oral-gut-microbial-network-disruption-in-hand-osteoarthritis-data-from-the-xiangya-osteoarthritis-study/
