Session Information
Date: Tuesday, October 28, 2025
Title: Abstracts: Metabolic & Crystal Arthropathies – Basic & Clinical Science (2585–2590)
Session Type: Abstract Session
Session Time: 2:15PM-2:30PM
Background/Purpose: Approximately 21% of US adults have hyperuricemia, the causal precursor for gout. Human cells do not degrade urate (due to uricase gene inactivation). However, a cluster of 7 bacterial genes in ~15-20% of commensal human gut purine degrading bacteria (PDB) encodes high capacity anaerobic uricolysis [1], distinct from strictly aerobic microbial uricase activity. Moreover, the majority of gut bacteria reside in the deeply anaerobic colonic lumen. By converting urate to xanthine, lactate, and short-chain fatty acids (SCFAs), PDB compensate for murine uricase knockout, thereby maintaining lower serum and cecal urate levels. Here, we seminally tested the association between gut PDB uricolytic genes and urate levels in humans.
Methods: We analyzed available blood and stool metagenomic (Illumina) data collected contemporaneously from men and women enrolled in three independent, prospective, nationwide cohort studies: Framingham Heart Study (FHS; Nf1129 men and women), Men’s Lifestyle Validation Study (MLVS; Nf296 men), and Mind Body Study (MBS; Nf207 women). The sequences of previously characterized urate-metabolizing genes (i.e., ygeX, ygeY, ygeW, ygfK, and ssnA) [1] were mapped to Uniref90 sequences and their corresponding species-level genomic bins, using HUMAnN and diamond [2-4]. We calculated the Spearman correlation between urate levels and each putative urate-metabolizing gene / Uniref90. These were visualized as undirected networks using the igraph and ggraph R packages.
Results: We identified 15 human gut-associated species-level genomic bins containing at least five urate-metabolizing genes in their pangenomes, corresponding to commensal gut species (Figure 1). These included Flavonifractor plautii, Enterocloster bolteae, and Ruminococcus torques. Figure 1 shows undirected associations between species-level genomic bins and their corresponding urate-metabolizing genes. In all, 12 of 48 identified urate-metabolizing genes significantly correlated with urate (spearman rho q < 0.1) in at least two studies, and 31 significantly correlated in at least one study (Figure 2). Additionally, 6 urate-metabolizing genes significantly correlated with urate only among participants in the MBS (i.e., women-only cohort) but not FHS or MLVS (Figure 2).
Conclusion: The abundance of putative urate-metabolizing genes is significantly correlated with urate in 3 human cohorts, and some may be sex-influenced. The commensal gut species identified may be particularly relevant to hyperuricemia and gout, including capacity for anti-inflammatory SCFA production (e.g., butyrate) and documented associations with other inflammatory conditions. These results, superimposed on known gut dysbiosis in gout [5], support actual modulation of urate by the gut microbiome in humans, and suggest a novel potential therapeutic target for prevention and treatment of gout.References:1. PMID: 375411972. PMID: 339447763. PMID: 368233564. PMID: 338282735. PMID: 39829115
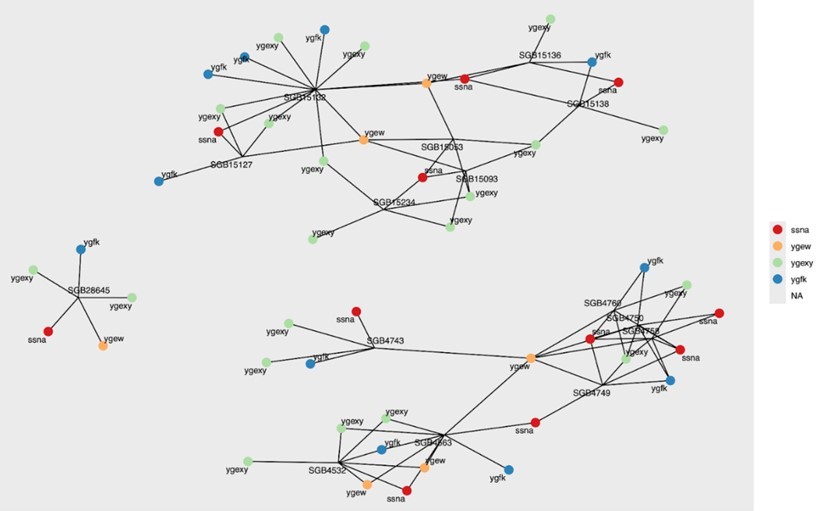 Figure 1. Undirected network diagram illustrating the associations between species-level genomic bins and corresponding urate-metabolizing genes.
Figure 1. Undirected network diagram illustrating the associations between species-level genomic bins and corresponding urate-metabolizing genes.
.jpg) Figure 2. Correlations between network Uniref90 genetic sequences and urate levels in each of the three cohorts. Symbols: *qval < 0.1, **qval < 0.05, ***qval < 0.01.
Figure 2. Correlations between network Uniref90 genetic sequences and urate levels in each of the three cohorts. Symbols: *qval < 0.1, **qval < 0.05, ***qval < 0.01.
To cite this abstract in AMA style:
Rai S, McCormick N, Morgan X, Nayor M, Terkeltaub R, Dodd D, Nazzal L, Li H, Curhan G, Huttenhower C, Choi H. Translating findings on urate-metabolizing bacterial genes and urate levels at the human population level: a gut microbiome analysis of three independent cohorts of men and women [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/translating-findings-on-urate-metabolizing-bacterial-genes-and-urate-levels-at-the-human-population-level-a-gut-microbiome-analysis-of-three-independent-cohorts-of-men-and-women/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/translating-findings-on-urate-metabolizing-bacterial-genes-and-urate-levels-at-the-human-population-level-a-gut-microbiome-analysis-of-three-independent-cohorts-of-men-and-women/
