Session Information
Session Type: Abstract Session
Session Time: 2:15PM-2:30PM
Background/Purpose: Timely diagnosis and clinically meaningful stratification of systemic lupus erythematosus (SLE) remain major unmet needs. Existing risk models rely on limited genetic or lifestyle predictors and are often built on non-representative cohorts. Phenotyping studies, meanwhile, typically use curated registries that fail to capture the real-world complexity of SLE presentations. The NIH All of Us program, a US-wide cohort, offers an unprecedented opportunity to integrate genetic risk scores (GRS), structured electronic health record (EHR) data, using unsupervised learning at scale to address both challenges.
Methods: Within All of Us (version 8), we identified SLE cases (≥2 ICD-10 codes on separate days) and matched controls by age, sex, and genetically inferred ancestry. Supervised models (logistic regression, random forest, XGBoost) were trained using 5-year pre-diagnosis features: GRS, medications, symptoms, reproductive, and self-reported lifestyle factors. Performance was assessed across time windows on a held-out test set.For phenotyping, we implemented two unsupervised clustering pipelines. Approach A used normalized immune, renal, and metabolic lab data during flare and non-flare periods. Approach B integrated post-diagnosis lab values, medication classes, and grouped conditions (6-18 months post-index). Dimensionality reduction (PCA, UMAP) preceded clustering (k-means, HDBSCAN). Clusters were interpreted based on lab profiles, treatment exposure, and organ-specific manifestations.
Results: 1526 SLE cases were matched to 3942 non-SLE controls. (Table 1) Prediction models yielded AUCs 0.61–0.64. GRS, joint pain, depression, and obesity were consistent predictors. Accuracy peaked near diagnosis (AUC 0.71), though at the cost of recall—highlighting the challenge of early signal detection.Clustering revealed distinct, interpretable phenotypes (Figures 1-2). A flare-associated cluster had concurrent dsDNA elevation, proteinuria, and transaminitis (renal/hepatic flare). A non-flare cluster featured persistent hypocomplementemia and hypertriglyceridemia, consistent with systemic inflammation and metabolic dysregulation. A 3rd group had high psychiatric and musculoskeletal burden with minimal serologic activity or immunosuppressant exposure—suggesting a care-intensive, serologically quiescent phenotype. Clusters demonstrated high clinical interpretability and mapped to established organ-domain patterns, with approach-specific silhouette scores from 0.27–0.49.
Conclusion: This is among the 1st studies to apply integrated machine learning across genomic and EHR and self-reported data to model both SLE risk and phenotypic heterogeneity. While predictive performance remains limited, phenotyping revealed actionable, domain-aligned subtypes with potential to inform prognosis, treatment, and clinical trial design. These findings highlight the promise of real-world, multimodal data to advance precision medicine in rheumatology.
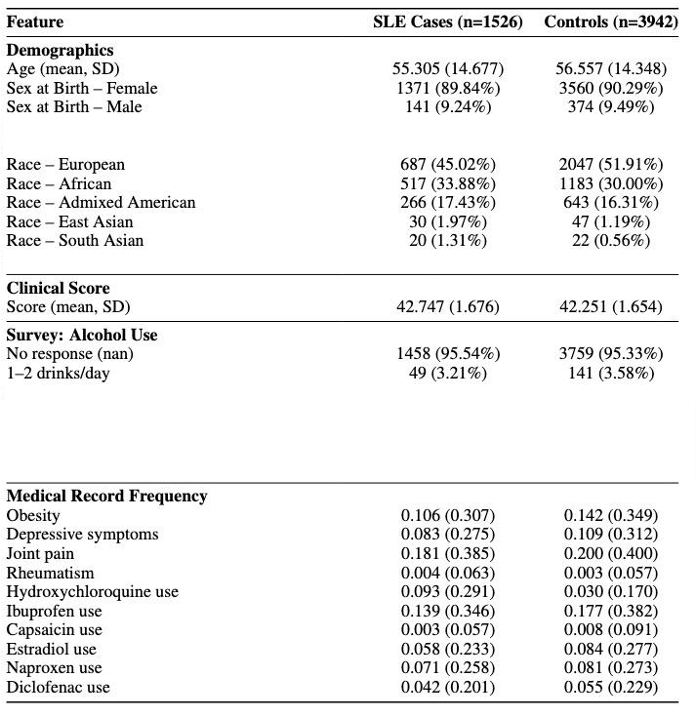 Table 1. Selected Demographic and Clinical Characteristics of the SLE Cases and their Matched Controls for 5 year – 1 year Pre-Diagnosis (SLE Diagnosis Index Date) in the All of Us Cohort, version 8
Table 1. Selected Demographic and Clinical Characteristics of the SLE Cases and their Matched Controls for 5 year – 1 year Pre-Diagnosis (SLE Diagnosis Index Date) in the All of Us Cohort, version 8
.jpg) Figure 1. SLE Patient Phenotypes for New Onset SLE Cases in the All of Us Program (version 8) using Genomic, EHR and Self-reported data (Flare-State Patients, Phenotyping Approach A)
Figure 1. SLE Patient Phenotypes for New Onset SLE Cases in the All of Us Program (version 8) using Genomic, EHR and Self-reported data (Flare-State Patients, Phenotyping Approach A)
.jpg) Figure 2. Top 10 Differentiating Features by Patient Type/Cluster (Phenotyping Approach B)
Figure 2. Top 10 Differentiating Features by Patient Type/Cluster (Phenotyping Approach B)
To cite this abstract in AMA style:
Sporn H, Parulekar-Martins R, Wang H, Yu X, Yee J, Kim Y, Cui J, Costenbader K. Multi-Modal Machine Learning Prediction and Phenotyping of Systemic Lupus Erythematosus Using Longitudinal EHR and Genomic Data from the All of Us Program [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/multi-modal-machine-learning-prediction-and-phenotyping-of-systemic-lupus-erythematosus-using-longitudinal-ehr-and-genomic-data-from-the-all-of-us-program/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/multi-modal-machine-learning-prediction-and-phenotyping-of-systemic-lupus-erythematosus-using-longitudinal-ehr-and-genomic-data-from-the-all-of-us-program/
