Session Information
Date: Tuesday, October 28, 2025
Title: (2470–2503) Systemic Sclerosis & Related Disorders – Clinical Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic sclerosis (SSc) is a clinically and biologically heterogeneous autoimmune disease characterized by multiorgan involvement, substantial morbidity, and high mortality. Traditional classification systems (based on the extent of cutaneous involvement or autoantibody status) fail to fully capture the complexity of disease expression and progression. There is a growing need for more precise patient stratification strategies that reflect underlying pathophysiology and can guide therapeutic decisions. Molecular approaches such as proteomic profiling may enable a shift from descriptive to mechanism-based classification frameworks in SSc.Our objective was to identify biologically and clinically distinct subgroups of SSc patients through unsupervised clustering of clinical and serological data, combined with high-throughput serum proteomic analysis.
Methods: Clinical and serological data from 402 patients in the multicenter PRECISESADS cohort were analyzed using k-means clustering to define SSc subtypes. Internal validity was assessed using statistical indices; external validation was performed in an independent Spanish cohort (n=213). Proteomic profiling was conducted on a representative subset of 154 patients using a targeted 92-protein OLINK® panel focused on organ damage. Functional validation was carried out by treating immortalized fibroblasts with serum from patients in each cluster, followed by RT-qPCR analysis of fibrotic and inflammatory gene expression.
Results: Two robust and clinically distinct clusters were identified and independently validated. Cluster 2 was associated with a more severe phenotype, including a higher prevalence of interstitial lung disease (59.7% vs. 14.5%), pulmonary arterial hypertension (30.9% vs. 9%), and musculoskeletal involvement. A detailed comparison of demographic and clinical characteristics between clusters is provided in Table 1.Proteomic analysis revealed 26 upregulated proteins in Cluster 2, including PDGFC, NOS3, and PRKAB1 (linked to fibrosis, endothelial injury, and metabolic stress (Figures 1A-D)). Compared to healthy donors, Cluster 2 showed greater proteomic dysregulation than Cluster 1 (Figures 2A-E), and pathway enrichment highlighted alterations in receptor tyrosine kinase signaling and immune activation. Functional assays confirmed that serum from Cluster 2 patients induced upregulation of profibrotic (PDGFRB, WNT3A) and proinflammatory (TNF, IL1B) genes in fibroblasts. Several proteins also correlated with clinical features such as pulmonary hypertension and esophageal dysmotility.
Conclusion: This integrative approach combining unsupervised clustering, proteomics, and functional validation identifies molecularly and clinically meaningful SSc subgroups. The findings support the use of circulating protein profiles to refine patient stratification and move toward personalized management in SSc.
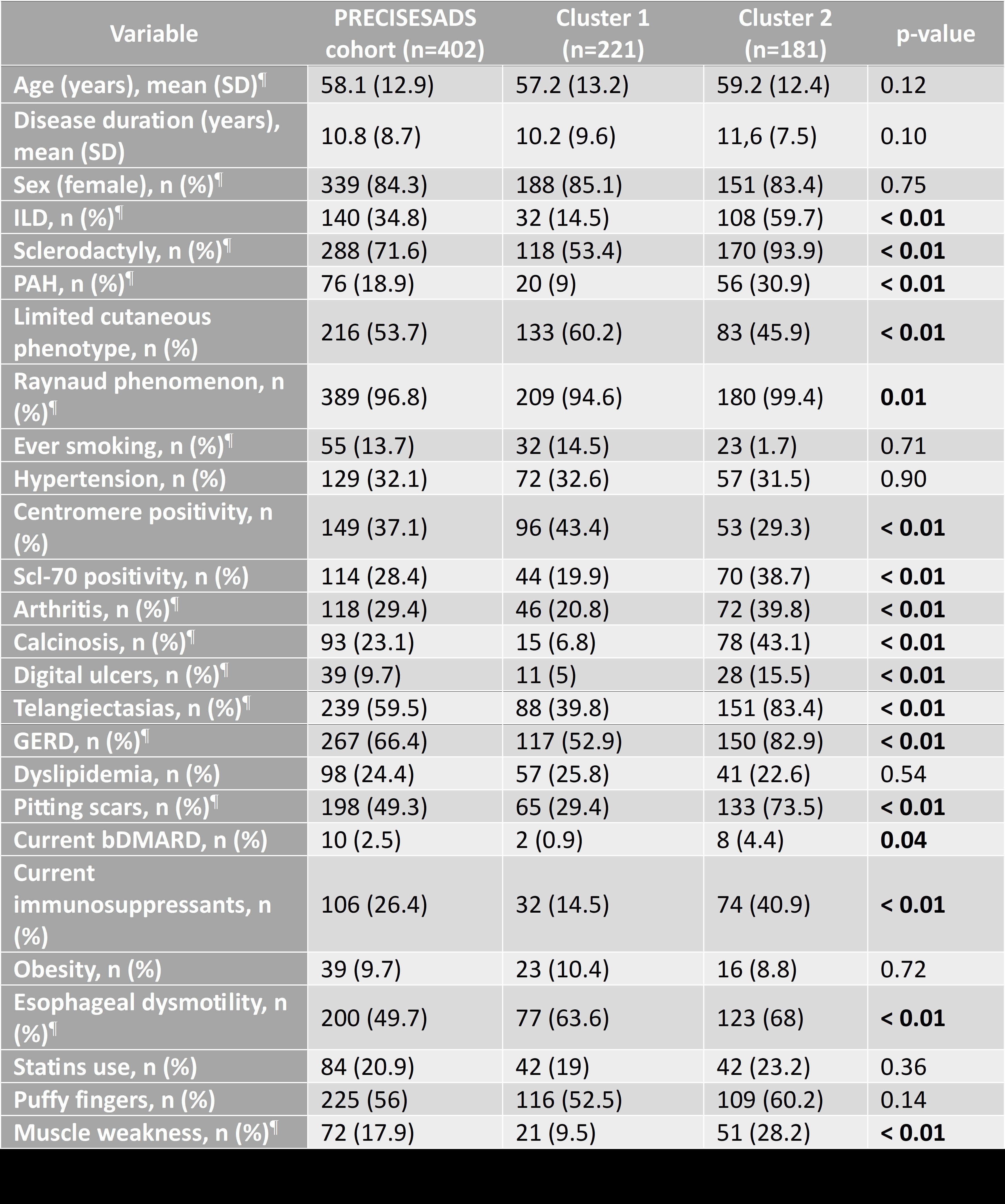 Table 1. Comparison of clinical and serological characteristics between the two identified clusters of SSc patients (PRECISESADS cohort). ¶: variables included in the clustering algorithm. ILD: interstitial lung disease; PAH: pulmonary arterial hypertension; GERD: gastroesophageal reflux disease; bDMARD: biologic disease-modifying antirheumatic drug.
Table 1. Comparison of clinical and serological characteristics between the two identified clusters of SSc patients (PRECISESADS cohort). ¶: variables included in the clustering algorithm. ILD: interstitial lung disease; PAH: pulmonary arterial hypertension; GERD: gastroesophageal reflux disease; bDMARD: biologic disease-modifying antirheumatic drug.
.jpg) Figure 1. Proteomic analysis of organ damage markers in SSc. (A) Principal component analysis (PCA) of SSc patient clusters based on organ damage-related proteomic markers. (B) Heatmap showing correlations among protein levels across all patients. (C) Volcano plot of differentially expressed proteins between SSc clusters. (D) Circulating levels of statistically significant proteins differentiating cluster 1 and cluster 2.
Figure 1. Proteomic analysis of organ damage markers in SSc. (A) Principal component analysis (PCA) of SSc patient clusters based on organ damage-related proteomic markers. (B) Heatmap showing correlations among protein levels across all patients. (C) Volcano plot of differentially expressed proteins between SSc clusters. (D) Circulating levels of statistically significant proteins differentiating cluster 1 and cluster 2.
.jpg) Figure 2. Comparative volcano plots and functional interaction network of differentially expressed proteins in SSc patient clusters vs healthy donors. (A) Volcano plot comparing protein expression between healthy controls and patients in Cluster 1. (B) Volcano plot showing the comparison between healthy controls and Cluster 2 patients, with a greater number of significantly upregulated proteins. (C) Venn diagram showing the overlap of differentially expressed proteins in clusters 1 and 2 compared to healthy donors. (D, E) Functional interaction network of the differentially expressed proteins compared to healthy controls, constructed using the STRING platform The analysis reveals a dysregulation of the Signaling by Receptor Tyrosine Kinases pathway (proteins in red) and signal transduction (proteins in purple).
Figure 2. Comparative volcano plots and functional interaction network of differentially expressed proteins in SSc patient clusters vs healthy donors. (A) Volcano plot comparing protein expression between healthy controls and patients in Cluster 1. (B) Volcano plot showing the comparison between healthy controls and Cluster 2 patients, with a greater number of significantly upregulated proteins. (C) Venn diagram showing the overlap of differentially expressed proteins in clusters 1 and 2 compared to healthy donors. (D, E) Functional interaction network of the differentially expressed proteins compared to healthy controls, constructed using the STRING platform The analysis reveals a dysregulation of the Signaling by Receptor Tyrosine Kinases pathway (proteins in red) and signal transduction (proteins in purple).
To cite this abstract in AMA style:
Dans Caballero S, Ortega-Castro R, López pedrera C, Escudero A, Vellón-García B, Pérez Sánchez C, López Medina C. Deciphering Systemic Sclerosis Phenotypes: A Novel Approach Using Clustering Algorithms and Proteomic Insights. Results from the PRECISESADS Study [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/deciphering-systemic-sclerosis-phenotypes-a-novel-approach-using-clustering-algorithms-and-proteomic-insights-results-from-the-precisesads-study/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deciphering-systemic-sclerosis-phenotypes-a-novel-approach-using-clustering-algorithms-and-proteomic-insights-results-from-the-precisesads-study/
