Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Juvenile dermatomyositis (JDM) is a multisystem vasculopathy and inflammatory myopathy characterized by proximal muscle weakness, distinct rash, and risk of long-term complications such as calcinosis, skin ulceration, and chronic disease. Although Type I and Type II Interferon signatures are well established, this alone does not explain the full pattern of disease; the basic biology of JDM remains poorly defined. Heterogeneity in outcomes necessitates a more fundamental understanding of disease mechanisms. Our objective is to understand genome-scale gene dysregulation in JDM through a multiomic study of single nuclei from treatment naïve muscle biopsies. We hypothesize that JDM etiology is mediated by molecular and cellular changes in the muscle and associated small blood vessels.
Methods: Muscle biopsies were collected from treatment naïve patients with JDM and congenital myopathy (CM). Biopsies were flash-frozen in liquid nitrogen and stored at -80°C (Cincinnati Children’s Hospital Medical Center (CCHMC) Pathology Core). Single nuclei isolation was performed (10x Genomics protocol with optimization performed on murine samples), followed by Chromium Next GEM Single Cell Multiome ATAC+ Gene Expression processing (CCHMC Single Cell Genomics Core). Sequencing data were analyzed through DNA and RNA read alignment and quantification using CellRanger. Ambient mRNA was removed, doublets identified, and low-quality nuclei removed using CellBender. Data were visualized in a two-dimensional space using dimensionality reduction and cluster identification using Seurat. Additional discovery of molecular pathway signatures was performed using CellHarmony.
Results: Single nuclei clustering revealed typical cell types consistent with previously demonstrated normal muscle tissue (Figure 1). Differentially expressed genes within individual cell types highlighted the known interferon signatures in multiple cell types. We also identified molecular signals consistent with dysfunction of vascular signaling pathways, especially between muscle and endothelial cells. There was marked upregulation of several inflammatory pathways that have not previously been described in JDM but have been identified in other inflammatory conditions (Figure 2).
Conclusion: We conducted a multiome study of single nuclei from treatment naïve muscle biopsies in JDM patients with a comparator group of congenital myopathy patients. While typical cell types of muscle are maintained, several pathways, in addition to a known interferon signature, demonstrate dysregulation, opening doors for further studies into the molecular dysfunction driving JDM pathology.
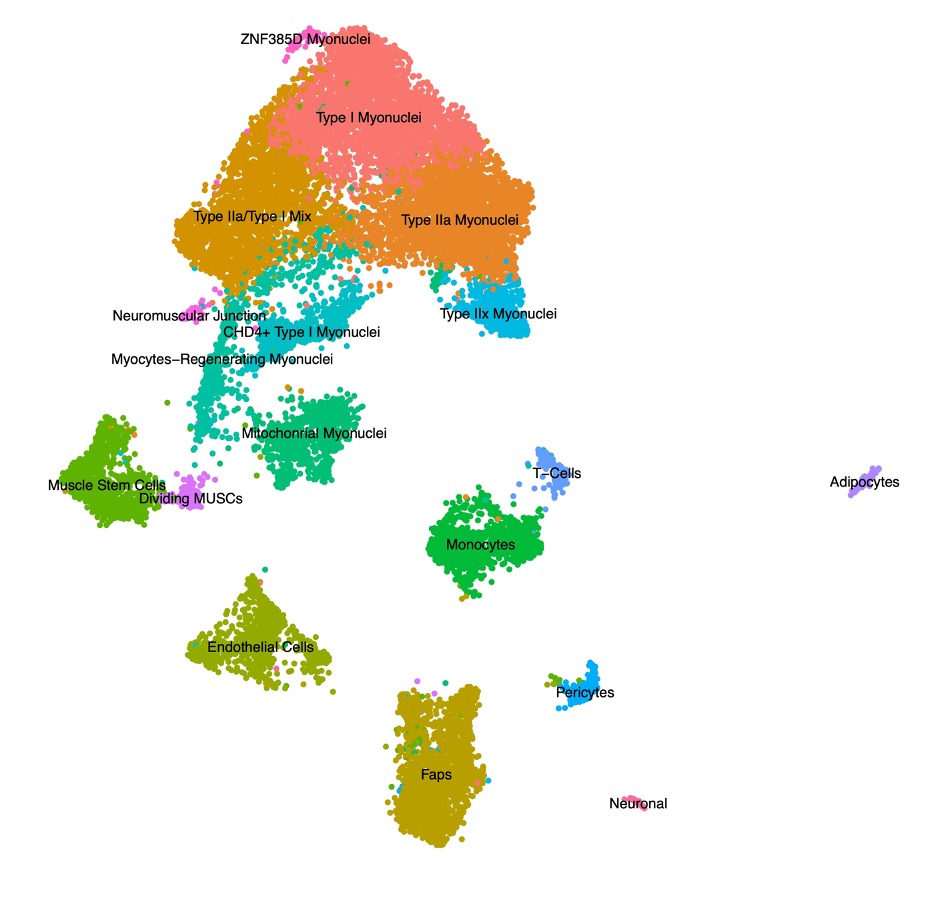 Figure 1. Uniform Manifold Approximation and Projection (UMAP) visualization represents sixteen color-coded nuclei clusters identified by tandem single nuclei RNA-sequencing and ATAC-sequencing. Data were generated from treatment naïve muscle biopsies from patients with juvenile dermatomyositis and congenital myopathy. FAPs = fibro-adipogenic precursors. MT = mitochondrial. NK = natural killer.
Figure 1. Uniform Manifold Approximation and Projection (UMAP) visualization represents sixteen color-coded nuclei clusters identified by tandem single nuclei RNA-sequencing and ATAC-sequencing. Data were generated from treatment naïve muscle biopsies from patients with juvenile dermatomyositis and congenital myopathy. FAPs = fibro-adipogenic precursors. MT = mitochondrial. NK = natural killer.
.jpg) Figure 2. Heatmap of differentially expressed genes by cell type (columns) highlighting significant changes in expression profiles across specific pathways (left side). Data generated by tandem single nuclei RNA-sequencing and ATAC-sequencing from treatment naïve muscle biopsies from patients with juvenile dermatomyositis and congenital myopathy. FAPs = fibro-adipogenic precursors.
Figure 2. Heatmap of differentially expressed genes by cell type (columns) highlighting significant changes in expression profiles across specific pathways (left side). Data generated by tandem single nuclei RNA-sequencing and ATAC-sequencing from treatment naïve muscle biopsies from patients with juvenile dermatomyositis and congenital myopathy. FAPs = fibro-adipogenic precursors.
To cite this abstract in AMA style:
O'Connor S, Swoboda C, Weirauch M, Zygmunt A, Millay D, Kottyan L. Single Nuclei Multiome of JDM Muscle Biopsies Reveals Novel Upregulation of Inflammatory and Vascular Pathways [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/single-nuclei-multiome-of-jdm-muscle-biopsies-reveals-novel-upregulation-of-inflammatory-and-vascular-pathways/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-nuclei-multiome-of-jdm-muscle-biopsies-reveals-novel-upregulation-of-inflammatory-and-vascular-pathways/
