Session Information
Date: Tuesday, October 28, 2025
Title: (1780–1808) Osteoarthritis & Joint Biology – Basic Science Poster
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: The knee menisci are essential in joint biomechanics while meniscus damage is a driver of knee osteoarthritis (OA) and an important source of knee pain. As early molecular and cellular changes mediating the onset of tissue damage and mechanisms of progression are poorly understood, there are no approaches to prevent, delay or stop progression of meniscus destruction. The goal of this study is to discover genes and mechanisms of meniscus aging and OA at early and late stages.
Methods: This study used human knees from organ donors, including young healthy donors and aged donors with macroscopically normal knee cartilage and menisci, aged donors with cartilage damage indicative of mild OA and patients with severe OA undergoing total knee arthroplasty (TKA) (A). The menisci were scored for histopathological changes (B). Bulk RNA-sequencing (RNA-seq) and single nuclei multiome were performed.
Results: Meniscus histopathology scores were normal in the young donor group (5.18±3.6) but also in the aged group (6.00±3.3) (B). The meniscus scores of the aged mild OA group were (9.14±2.7) and of the TKA group were 13.0±2.7 (B). RNA-seq detected only three DEGs in the menisci from aged normal donors as compared to the young normal. The aged OA group showed some separation from the young normal group on a principal component analysis (PCA) plot (C) and had 84 downregulated genes (DRGs) and 413 upregulated genes (URGs) (D). Metascape analysis revealed biological processes (BPs) regulated by the DRGs including macroautophagy, Endoplasmic-reticulum-associated protein degradation (ERAD), protein folding and NABA core matrisome (E, F). The URGs were also enriched for BPs related to extracellular matrix (ECM) and sensory system remodeling (E). The TKA group showed clear separation from the young normal group on PCA plot (G) and had 577 DRGs and 2680 URGs (H). The DRGs were enriched for metabolic pathways and protective BPs related to response to amino acid deficiency and antimicrobial defense (I). The URGs were strongly enriched for matrisome and ECM and for neuronal system, synaptic transmission and sensory organ development (I). Among the URGs were genes related to pain, including NGF and ANK3 (J), and genes encoding voltage-gated sodium channels and transient receptor potential cation channels (K). In multiome analysis, several cell populations were significantly enriched in TKA compared to normal (clusters 0, 10 and 16). Cluster 10 had 1554 differentially expressed genes with enrichment for the OA-promoting genes, pathways related to calcium signaling, ECM, chondrocyte/bone differentiation and mitochondrial dysfunction. NFATC2 and NFATC1 were differentially expressed in cluster 10 in TKA compared to normal and had enhanced regulon activity.
Conclusion: Early or mild stages of OA are primarily characterized by disruption of cellular homeostasis and defense mechanisms and a relatively low level of ECM process activation. These processes are more enriched in late-stage OA which also has a marked enhancement of processes related to neuronal remodeling and pain. A pathogenic cell population that is expanded in OA and is a promising new therapeutic target.
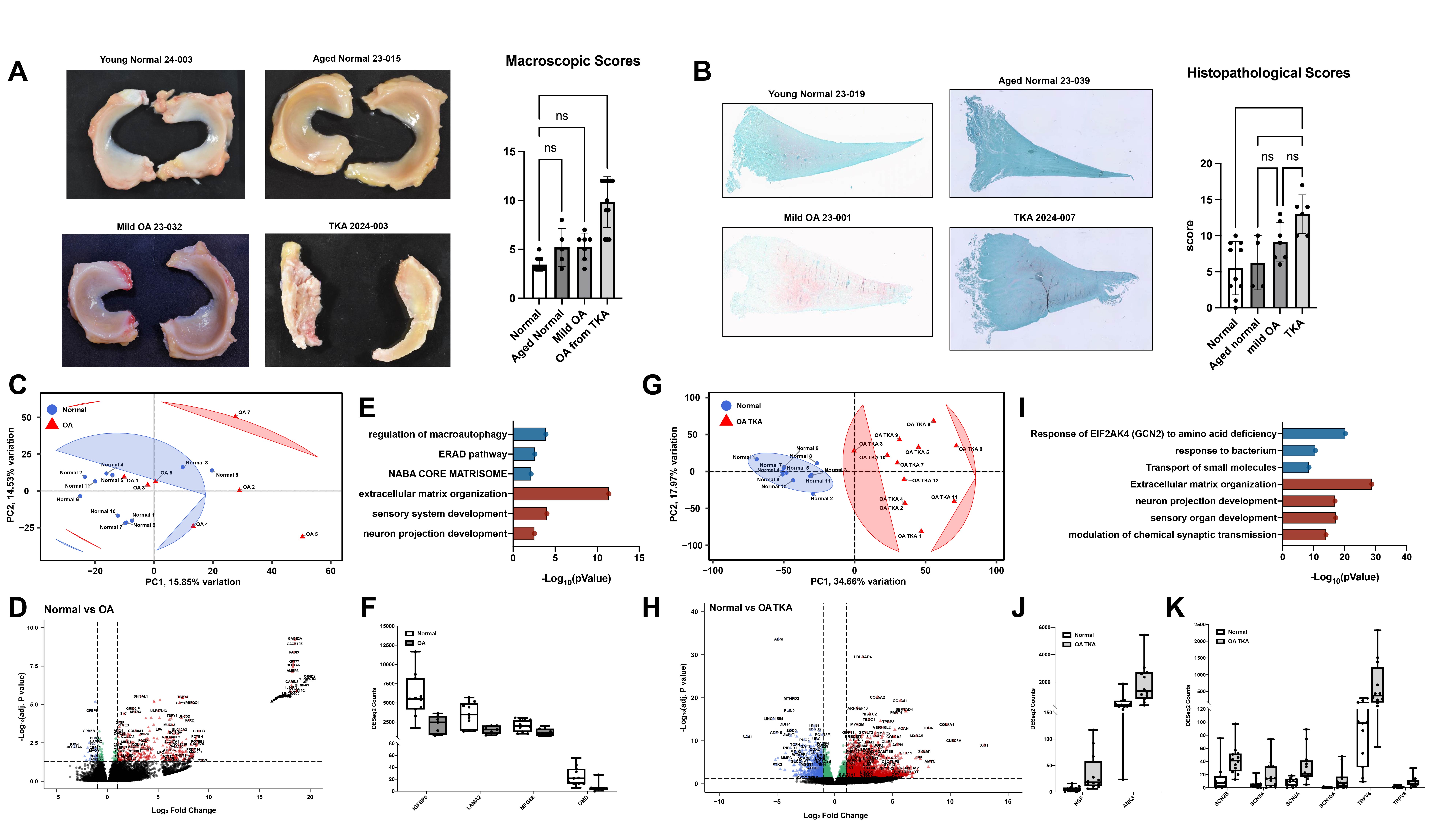 (A) Representative macroscopic images (left) and quantification of macroscopic scores (right) for young normal, aged normal, mild OA and TKA menisci. (B) Representative histologic images (left) and quantification of histological scores (right) for young normal, aged normal, mild OA and TKA menisci. (C) PCA plot showing separation between young normal and mild OA groups. (D) Volcano plot showing the differentially expressed genes (adj p < 0.05 and baseMean > 1). (E) Metascape functional enrichment analysis showing enrichment of biological processes in young normal (blue) and mild OA (red). (F) Box plots showing DESeq2 counts of several differentially expressed genes between young normal and mild OA menisci. (G) PCA plot showing separation between young normal and TKA groups. (H) Volcano plot showing the differentially expressed genes (adj p < 0.05 and log2FC > |1|) . (I) Metascape functional enrichment analysis showing enrichment of biological processes in young normal (blue) and TKA (red). (J, K) Box plots showing DESeq2 counts of several differentially expressed genes between young normal and TKA menisci.
(A) Representative macroscopic images (left) and quantification of macroscopic scores (right) for young normal, aged normal, mild OA and TKA menisci. (B) Representative histologic images (left) and quantification of histological scores (right) for young normal, aged normal, mild OA and TKA menisci. (C) PCA plot showing separation between young normal and mild OA groups. (D) Volcano plot showing the differentially expressed genes (adj p < 0.05 and baseMean > 1). (E) Metascape functional enrichment analysis showing enrichment of biological processes in young normal (blue) and mild OA (red). (F) Box plots showing DESeq2 counts of several differentially expressed genes between young normal and mild OA menisci. (G) PCA plot showing separation between young normal and TKA groups. (H) Volcano plot showing the differentially expressed genes (adj p < 0.05 and log2FC > |1|) . (I) Metascape functional enrichment analysis showing enrichment of biological processes in young normal (blue) and TKA (red). (J, K) Box plots showing DESeq2 counts of several differentially expressed genes between young normal and TKA menisci.
To cite this abstract in AMA style:
Sakamoto T, Olmer M, Kenvisay c, Miller R, Malfait A, D'Lima D, Bugbee W, Swahn H, Lotz M. Human meniscus histopathological and transcriptomic changes at early and advanced stages of knee osteoarthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/human-meniscus-histopathological-and-transcriptomic-changes-at-early-and-advanced-stages-of-knee-osteoarthritis-2/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/human-meniscus-histopathological-and-transcriptomic-changes-at-early-and-advanced-stages-of-knee-osteoarthritis-2/
