Session Information
Date: Monday, October 27, 2025
Title: (1191–1220) Muscle Biology, Myositis & Myopathies – Basic & Clinical Science Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Idiopathic inflammatory myopathies (IIMs) are rare autoimmune muscle disorders with complex genetic underpinnings. Most genetic studies have focused on limited populations. This study explores genetic risk factors in Indian IIM patients, integrating clinical, serological, and histopathological features to enhance understanding of its pathogenesis.
Methods: Genomic DNA was isolated from EDTA blood and libraries were prepared using the SureSelect XT HS2 protocol and sequenced on the Illumina NextSeq 2000 (2×150 bp). Variants were prioritized based on quality, frequency, predicted impact, and pathway relevance then selected variants were validated by Sanger sequencing using custom-designed primers and standard PCR. Frequently mutated genes were assessed for phenotype associations.
Results: A total of 91 prioritized variants were identified across 86 IIM cases, with mutations present in 68.6% (59/86). Of these, 4.4% were classified as pathogenic, 14.3% as likely pathogenic, and 81.3% as variants of uncertain significance (VUS). Most were nonsynonymous SNVs (80.2%), followed by frameshift deletions (5.5%), stopgain mutations (4.4%), and others. Notably, 25.3% were novel variants. Seventeen genes harbored ≥2 variants or at least one pathogenic/likely pathogenic variant, with five genes (CFTR, COL1A1, HBB, LRP5, and WDFY4) meeting both criteria for further evaluation. Among these, LRP5 carried five nonsynonymous SNVs across IIM subgroups. Domain mapping revealed key mutations within or adjacent to EGF-like and LDL-receptor domains (Figure 1). LRP5 mutations were significantly associated with proximal lower limb weakness (p=0.0372), anti-Ro52 (p=0.0369), and anti-TIF1γ autoantibodies (p=0.0156) (Table 1), suggesting a potential genotype-phenotype correlation.
Conclusion: This study identifies key genetic variants in Indian IIM patients, highlighting LRP5 as a potential marker linked to distinct clinical and serological features, advancing understanding of IIM pathogenesis.
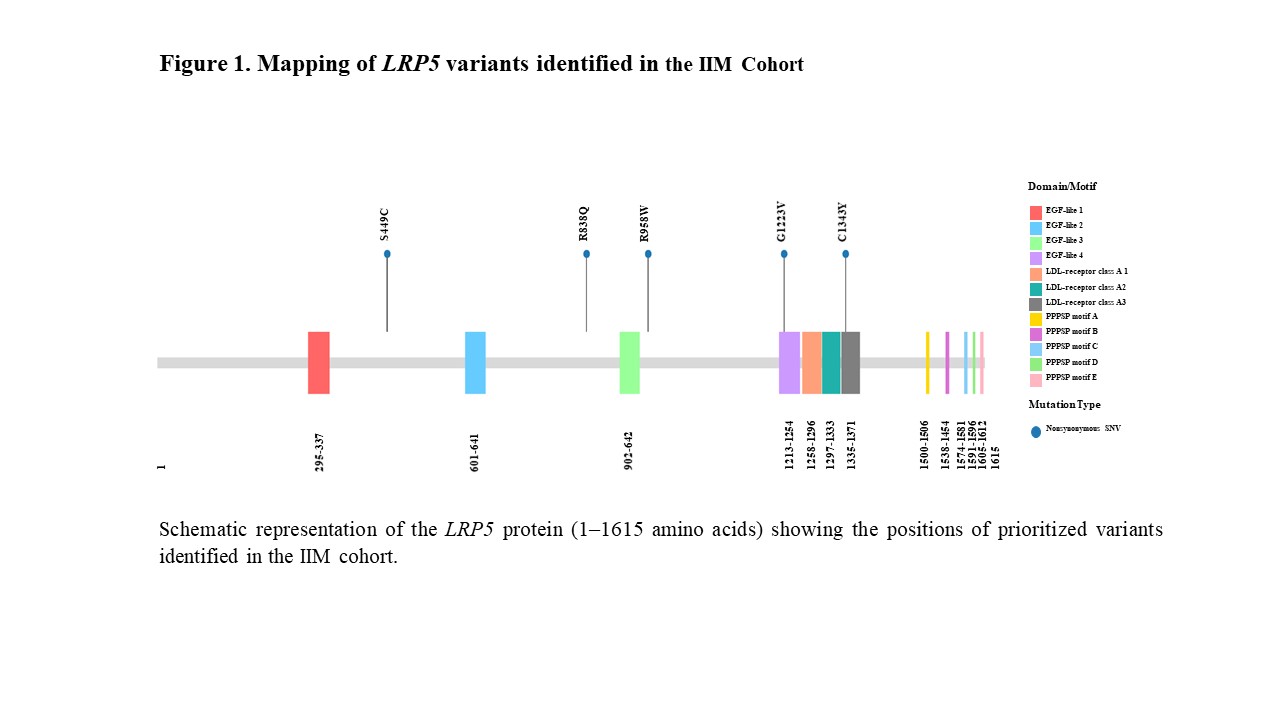 Figure 1. Mapping of LRP5 variants identified in the IIM Cohort
Figure 1. Mapping of LRP5 variants identified in the IIM Cohort
.jpg) Table 1. Comparison of characteristics of IIM Patients With and Without LRP5 Variants: This table presents a univariate comparison between idiopathic inflammatory myopathy (IIM) patients carrying prioritized LRP5 variants (n = 5) and those without such variants (n = 81). Clinical, histopathological, serological, and diagnostic variables are shown as counts with corresponding percentages. Statistical significance was assessed using Fisher’s exact test. Results are expressed as p-values with corresponding odds ratios (OR) and 95% confidence intervals (CI) in parentheses.
Table 1. Comparison of characteristics of IIM Patients With and Without LRP5 Variants: This table presents a univariate comparison between idiopathic inflammatory myopathy (IIM) patients carrying prioritized LRP5 variants (n = 5) and those without such variants (n = 81). Clinical, histopathological, serological, and diagnostic variables are shown as counts with corresponding percentages. Statistical significance was assessed using Fisher’s exact test. Results are expressed as p-values with corresponding odds ratios (OR) and 95% confidence intervals (CI) in parentheses.
To cite this abstract in AMA style:
Jassal B, Dhall A, Faruq M, Bhadu D, Kumar U, Sharma M. Genetic Profiling of Idiopathic Inflammatory Myopathies in Indian Patients Reveals Genotype-Phenotype Correlation [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/genetic-profiling-of-idiopathic-inflammatory-myopathies-in-indian-patients-reveals-genotype-phenotype-correlation/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-profiling-of-idiopathic-inflammatory-myopathies-in-indian-patients-reveals-genotype-phenotype-correlation/
